|
|
|
|
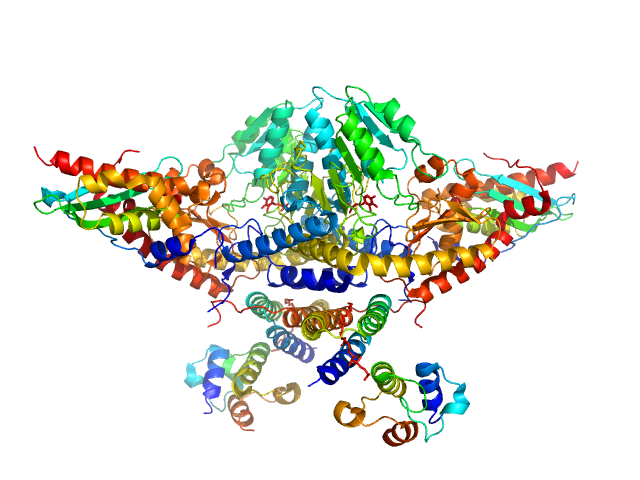
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 May 22
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
3.9 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
|
|
|
|
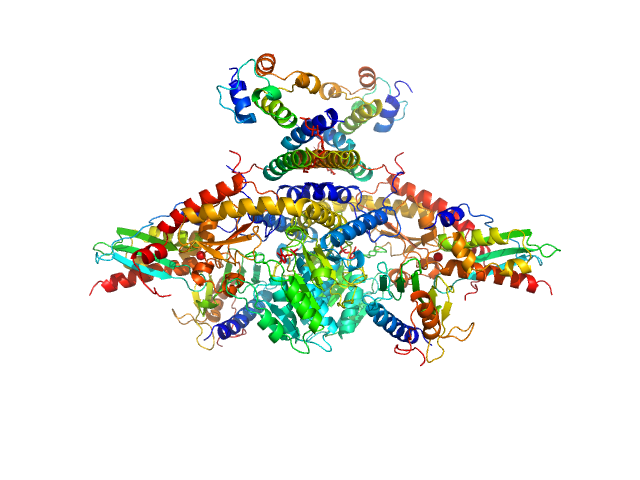
|
| Sample: |
Cysteine desulfurase, mitochondrial dimer, 90 kDa Homo sapiens protein
LYR motif-containing protein 4 dimer, 23 kDa Homo sapiens protein
Acyl carrier protein dimer, 22 kDa Escherichia coli protein
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial dimer, 29 kDa Homo sapiens protein
Frataxin, mitochondrial dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at Bruker Nanostar, NMRFAM on 2017 Apr 18
|
Architectural Features of Human Mitochondrial Cysteine Desulfurase Complexes from Crosslinking Mass Spectrometry and Small-Angle X-Ray Scattering.
Structure 26(8):1127-1136.e4 (2018)
Cai K, Frederick RO, Dashti H, Markley JL
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
287 |
nm3 |
|
|
|
|
|
|
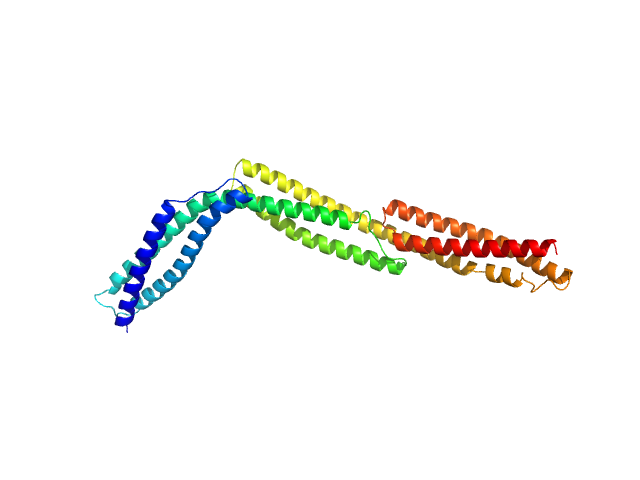
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.7 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
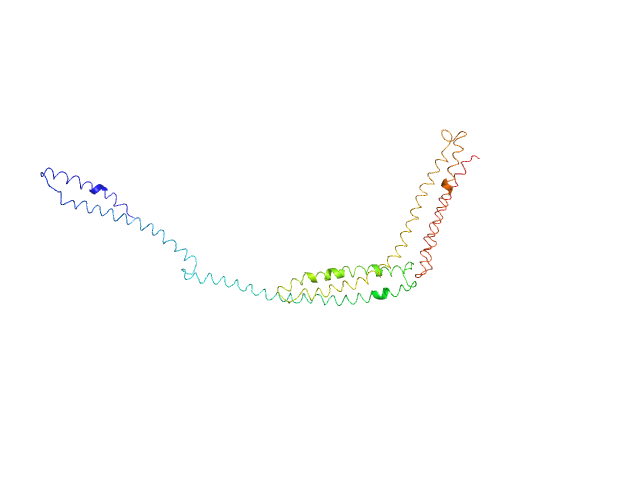
|
| Sample: |
R1-3 human dystrophin fragment monomer, 39 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-d11, 150 mM NaCl, 0.1 mM EDTA-d16, in 100% D2O, pD 7.5, pH: 7.1 |
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2016 Nov 7
|
Human Dystrophin Structural Changes upon Binding to Anionic Membrane Lipids.
Biophys J 115(7):1231-1239 (2018)
Dos Santos Morais R, Delalande O, Pérez J, Mias-Lucquin D, Lagarrigue M, Martel A, Molza AE, Chéron A, Raguénès-Nicol C, Chenuel T, Bondon A, Appavou MS, Le Rumeur E, Combet S, Hubert JF
|
| RgGuinier |
6.2 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Anti-CD32b Antibody Clone 6G08 Antibody Binding Fragment monomer, 46 kDa Homo sapiens protein
|
| Buffer: |
50mM HEPES, 150mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 Dec 5
|
Evaluating Anti-CD32b F(ab) Conformation Using Molecular Dynamics and Small-Angle X-Ray Scattering.
Biophys J 115(2):289-299 (2018)
Sutton EJ, Bradshaw RT, Orr CM, Frendéus B, Larsson G, Teige I, Cragg MS, Tews I, Essex JW
|
| RgGuinier |
2.6 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
68 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Small glutamine-rich tetratricopeptide repeat-containing protein alpha full length dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 5
|
Structural complexity of the co-chaperone SGTA: a conserved C-terminal region is implicated in dimerization and substrate quality control.
BMC Biol 16(1):76 (2018)
Martínez-Lumbreras S, Krysztofinska EM, Thapaliya A, Spilotros A, Matak-Vinkovic D, Salvadori E, Roboti P, Nyathi Y, Muench JH, Roessler MM, Svergun DI, High S, Isaacson RL
|
|
|
|
|
|
|
|
| Sample: |
Small glutamine-rich tetratricopeptide repeat-containing protein alpha Nterminal-TPR domains dimer, 47 kDa Homo sapiens protein
|
| Buffer: |
10 mM potassium phosphate, 100 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 5
|
Structural complexity of the co-chaperone SGTA: a conserved C-terminal region is implicated in dimerization and substrate quality control.
BMC Biol 16(1):76 (2018)
Martínez-Lumbreras S, Krysztofinska EM, Thapaliya A, Spilotros A, Matak-Vinkovic D, Salvadori E, Roboti P, Nyathi Y, Muench JH, Roessler MM, Svergun DI, High S, Isaacson RL
|
|
|
|
|
|
|

|
| Sample: |
Designed Ankyrin Repeat Protein D1 monomer, 18 kDa synthetic construct protein
Kinesin-like protein KIF2A monomer, 48 kDa Homo sapiens protein
Tubulin alpha-1B chain dimer, 100 kDa Bos taurus protein
Tubulin beta-2B chain dimer, 100 kDa Bos taurus protein
|
| Buffer: |
HEPES 20 mM, MgCl2 1mM, NaCl 150mM, pH: 7.2 |
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2016 May 16
|
Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate.
Nat Commun 9(1):2628 (2018)
Trofimova D, Paydar M, Zara A, Talje L, Kwok BH, Allingham JS
|
| RgGuinier |
5.4 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
374 |
nm3 |
|
|
|
|
|
|
|
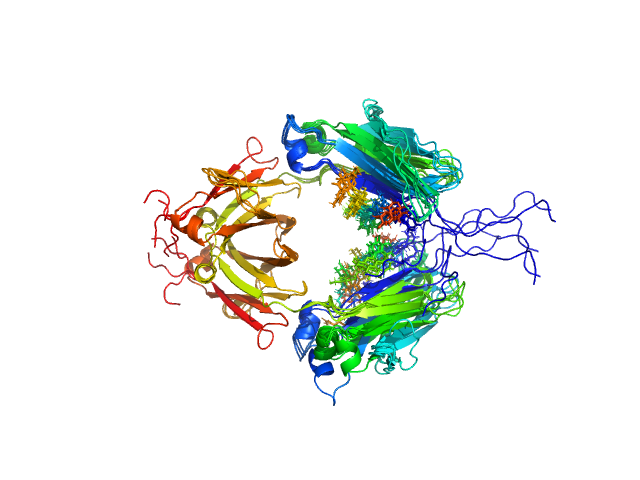
| Sample: |
Immunoglobulin heavy constant gamma 1 dimer, 53 kDa Homo sapiens protein
|
| Buffer: |
20mM HEPES, 50mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2016 Feb 17
|
Conformational Plasticity of the Immunoglobulin Fc Domain in Solution.
Structure 26(7):1007-1014.e2 (2018)
Remesh SG, Armstrong AA, Mahan AD, Luo J, Hammel M
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|