|
|
|
|
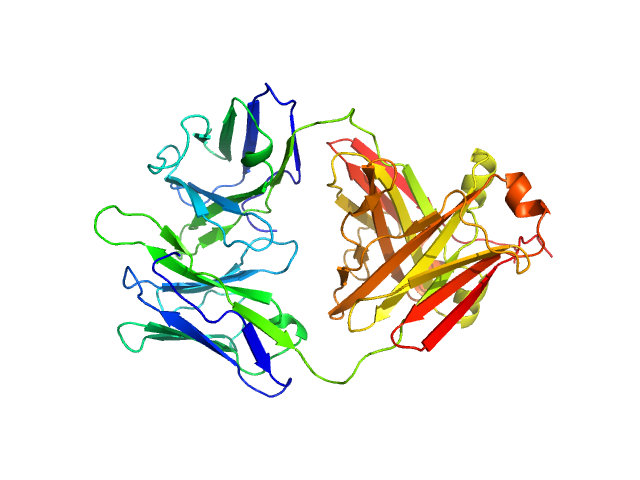
|
| Sample: |
Immunoglobulin light chain H7 dimer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 12
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
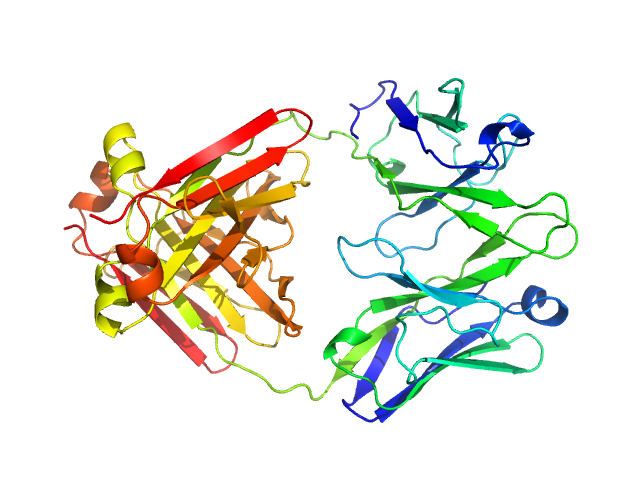
|
| Sample: |
Immunoglobulin light chain H18 dimer, 43 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2021 Jul 2
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
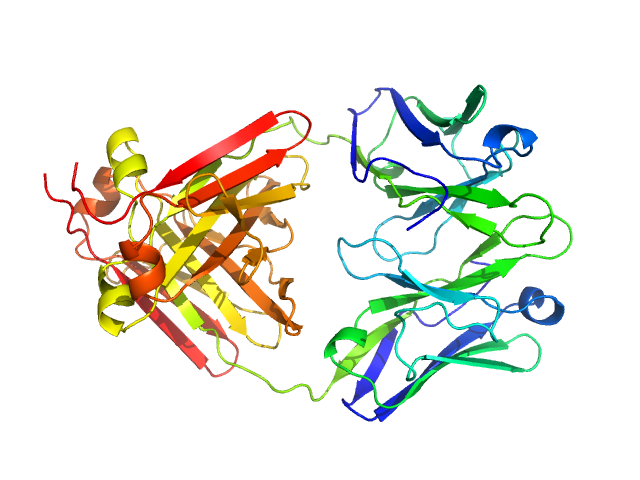
|
| Sample: |
Immunoglobulin light chain M7 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 12
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
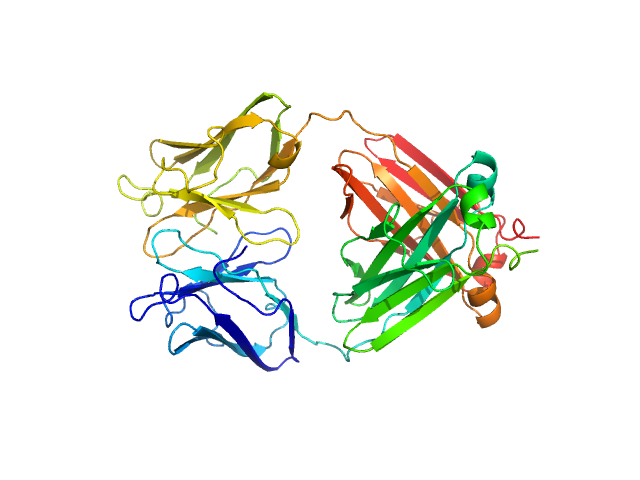
|
| Sample: |
Immunoglobulin light chain M10 dimer, 45 kDa Homo sapiens protein
|
| Buffer: |
20 mM TrisHCL, 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Mar 17
|
A conformational fingerprint for amyloidogenic light chains
eLife 13 (2025)
Paissoni C, Puri S, Broggini L, Sriramoju M, Maritan M, Russo R, Speranzini V, Ballabio F, Nuvolone M, Merlini G, Palladini G, Hsu S, Ricagno S, Camilloni C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-dependent DNA helicase Q5, 109 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2023 Mar 29
|
Mechanisms of transcription attenuation and condensation of RNA polymerase II by RECQ5
Tomas Klumpler
|
| RgGuinier |
6.5 |
nm |
| Dmax |
19.5 |
nm |
| VolumePorod |
310 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-dependent DNA helicase Q5, 109 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2023 Mar 29
|
Mechanisms of transcription attenuation and condensation of RNA polymerase II by RECQ5
Tomas Klumpler
|
| RgGuinier |
6.4 |
nm |
| Dmax |
20.2 |
nm |
| VolumePorod |
288 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-dependent DNA helicase Q5, 21 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2023 Apr 19
|
Mechanisms of transcription attenuation and condensation of RNA polymerase II by RECQ5
Tomas Klumpler
|
| RgGuinier |
3.7 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
73 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-dependent DNA helicase Q5, 109 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2023 Apr 19
|
Mechanisms of transcription attenuation and condensation of RNA polymerase II by RECQ5
Tomas Klumpler
|
| RgGuinier |
3.9 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
91 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-dependent DNA helicase Q5, 109 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2023 Apr 19
|
Mechanisms of transcription attenuation and condensation of RNA polymerase II by RECQ5
Tomas Klumpler
|
| RgGuinier |
4.3 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
116 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ATP-dependent DNA helicase Q5, 109 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2023 Apr 19
|
Mechanisms of transcription attenuation and condensation of RNA polymerase II by RECQ5
Tomas Klumpler
|
| RgGuinier |
4.6 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
102 |
nm3 |
|
|