|
|
|
|
|
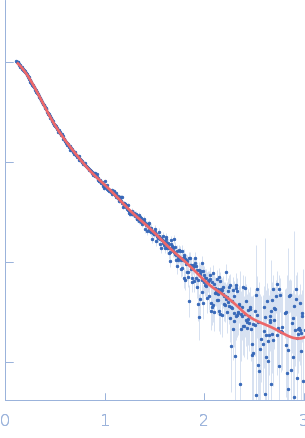
| Sample: |
ATP-dependent DNA helicase Q5, 109 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 200 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2023 Apr 19
|
Mechanisms of transcription attenuation and condensation of RNA polymerase II by RECQ5
Tomas Klumpler
|
| RgGuinier |
5.7 |
nm |
| Dmax |
16.2 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
|
|
|
|
|
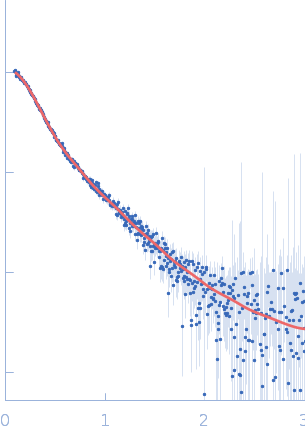
| Sample: |
Endophilin-B1 dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
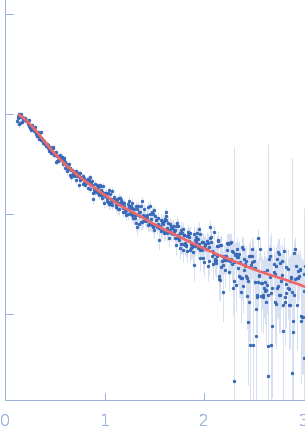
| Sample: |
Endophilin-B1 dimer, 82 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
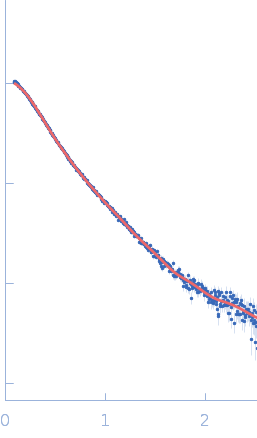
| Sample: |
Endophilin-B1 monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.3 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endophilin-B1 (Δ307-360) dimer, 68 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM TCEP, 0.5 mM DTT, pH: 8.1 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2024 Mar 10
|
Peripheral membrane protein endophilin B1 probes, perturbs and permeabilizes lipid bilayers
Communications Biology 8(1) (2025)
Thorlacius A, Rulev M, Sundberg O, Sundborger-Lunna A
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.2 |
nm |
| VolumePorod |
149 |
nm3 |
|
|
|
|
|
|
|
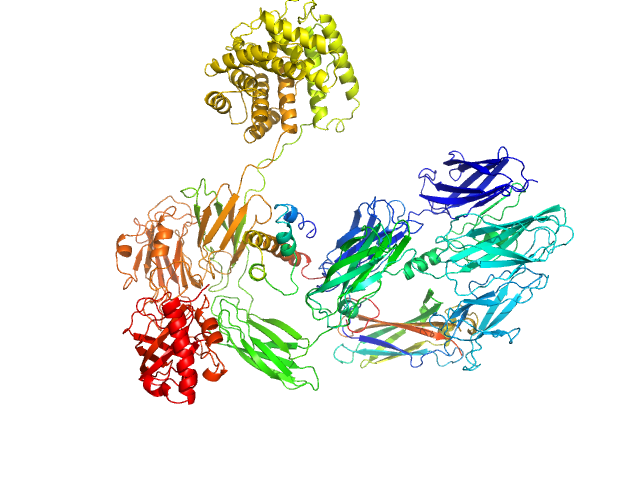
| Sample: |
Complement C3 (Δ668-671) monomer, 187 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES pH 6.0, 200 mM NaCl, pH: 6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 7
|
Cryo-EM analysis of complement C3 reveals a reversible major opening of the macroglobulin ring.
Nat Struct Mol Biol (2025)
Gadeberg TAF, Jørgensen MH, Olesen HG, Lorentzen J, Harwood SL, Almeida AV, Fruergaard MU, Jensen RK, Kanis P, Pedersen H, Tranchant E, Petersen SV, Thøgersen IB, Kragelund BB, Lyons JA, Enghild JJ, Andersen GR
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.8 |
nm |
| VolumePorod |
357 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
3.1 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 monomer, 16 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 Oct 26
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
52 |
nm3 |
|
|