|
|
|
|
|
| Sample: |
Mitochondrial intermembrane space import and assembly protein 40 dimer, 12 kDa Homo sapiens protein
Apoptosis-inducing factor 1, mitochondrial dimer, 113 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Nov 28
|
NADH-bound AIF activates the mitochondrial CHCHD4/MIA40 chaperone by a substrate-mimicry mechanism.
EMBO J (2025)
Brosey CA, Shen R, Tainer JA
|
| RgGuinier |
3.9 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
289 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor (mutant S118A) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.2 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
71 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor (mutant S118D) monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 150 mM NaCl, 0.5 mM EDTA, 0.1 mM PMSF, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2019 Jul 21
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.5 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Estrogen receptor monomer, 20 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate, 50 mM NaCl, 0.05 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2024 Jul 17
|
The sequence–structure–function relationship of intrinsic ERα disorder
Nature (2025)
Du Z, Wang H, Luo S, Yun Z, Wu C, Yang W, Buck M, Zheng W, Hansen A, Kao H, Yang S
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
70 |
nm3 |
|
|
|
|
|
|
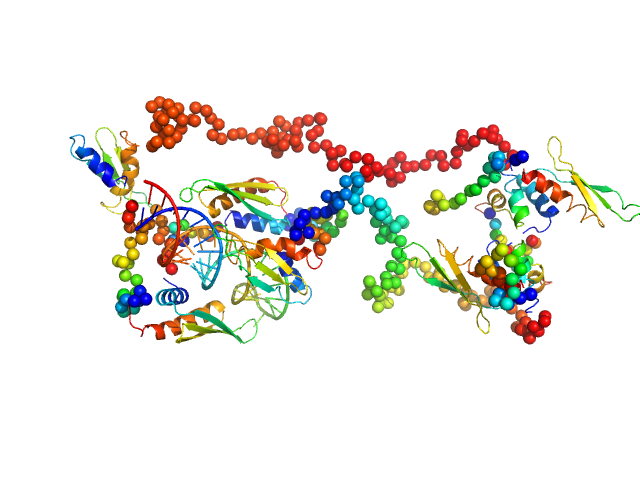
|
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 (Δ1-177) dimer, 89 kDa Homo sapiens protein
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2020 May 4
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|
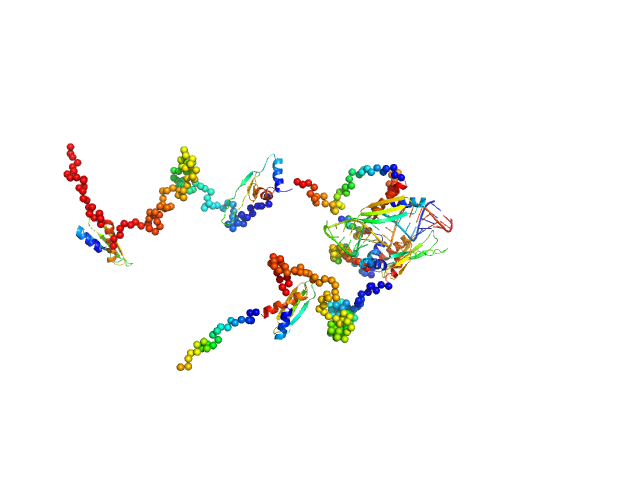
|
| Sample: |
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 with truncated RNA-binding domain 2 and truncated Staufen-swapping (ΔSSM) dimer, 81 kDa Homo sapiens protein
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2024 Jan 12
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 11
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
5.5 |
nm |
| Dmax |
25.4 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 160 mM NaCl, 0.5 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 11
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
4.7 |
nm |
| Dmax |
21.3 |
nm |
| VolumePorod |
197 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 (L24A) monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 160 mM NaCl, 0.5 mM TCEP pH 7.5, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Dec 9
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
127 |
nm3 |
|
|