|
|
|
|
|
| Sample: |
Importin subunit alpha-1 monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline containing 2 M urea, 0.3 M KCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Jul 1
|
A genetically encoded anomalous SAXS ruler to probe the dimensions of intrinsically disordered proteins.
Proc Natl Acad Sci U S A 121(50):e2415220121 (2024)
Yu M, Gruzinov AY, Ruan H, Scheidt T, Chowdhury A, Giofrè S, Mohammed ASA, Caria J, Sauter PF, Svergun DI, Lemke EA
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Importin-beta-binding domain of importin subunit alpha-1 labelled with unnatural amino acid diBrK monomer, 8 kDa Homo sapiens protein
|
| Buffer: |
phosphate buffered saline containing 6 M urea, 0.3 M KCl, 5 mM DTT, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Dec 19
|
A genetically encoded anomalous SAXS ruler to probe the dimensions of intrinsically disordered proteins.
Proc Natl Acad Sci U S A 121(50):e2415220121 (2024)
Yu M, Gruzinov AY, Ruan H, Scheidt T, Chowdhury A, Giofrè S, Mohammed ASA, Caria J, Sauter PF, Svergun DI, Lemke EA
|
| RgGuinier |
3.0 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
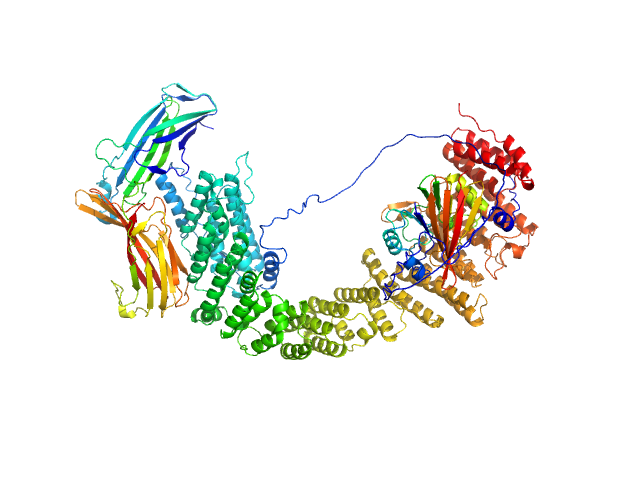
| Sample: |
VPS35 endosomal protein-sorting factor-like monomer, 110 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 29 monomer, 21 kDa Homo sapiens protein
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 200 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Apr 28
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
|
|
|
|
|
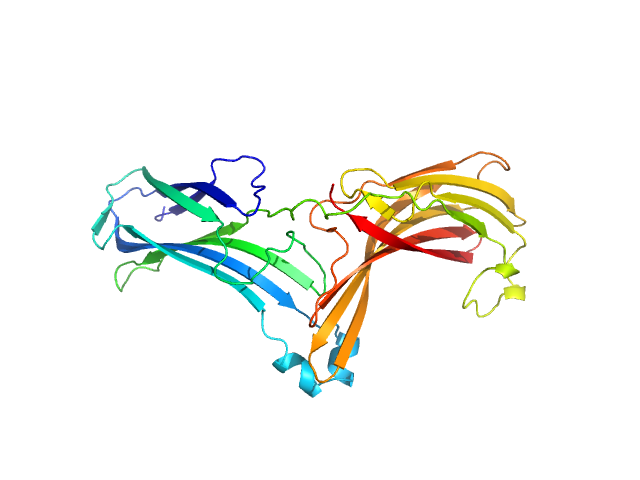
| Sample: |
Vacuolar protein sorting-associated protein 26C monomer, 33 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Apr 22
|
Selective cargo and membrane recognition by SNX17 regulates its interaction with Retriever
EMBO Reports (2024)
Martín-González A, Méndez-Guzmán I, Zabala-Zearreta M, Quintanilla A, García-López A, Martínez-Lombardía E, Albesa-Jové D, Acosta J, Lucas M
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 688-817, i.e. dsRBD3-long) dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
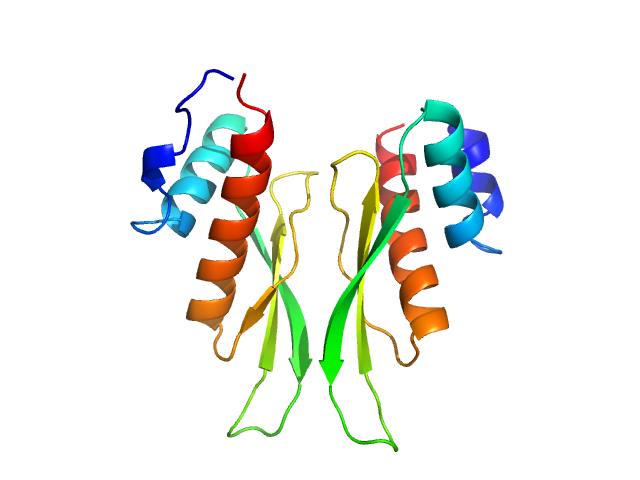
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 708-801, i.e. dsRBD3-mid) dimer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 716-797, i.e. dsRBD3-short) dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (wild-type, residues 708-801, i.e. ADAR1-dsRBD3) dimer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (interface mutant, residues 708-801, i.e. ADAR1-dsRBD3 interface mutant) monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Chimeric construct between the third dsRBD of human ADAR1 and the second dsRBD of Xenopus Xlrbpa (chimeric construct, residues 708-801, i.e. Chimeric ADAR1-ds3/Xlrbpa-ds2) monomer, 13 kDa Homo sapiens / … protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|