|
|
|
|
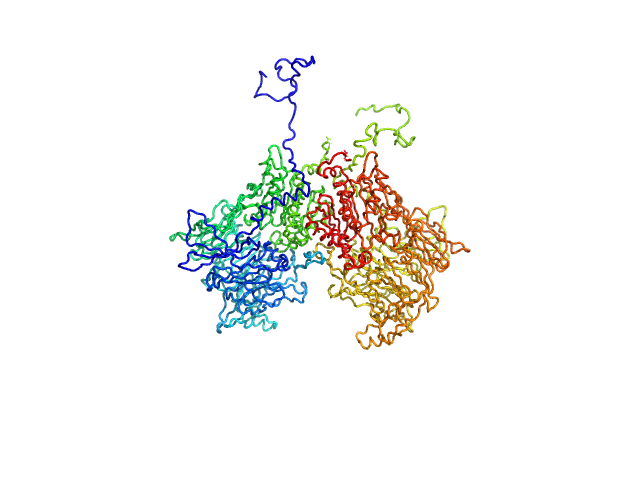
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.6 |
nm |
| VolumePorod |
271 |
nm3 |
|
|
|
|
|
|
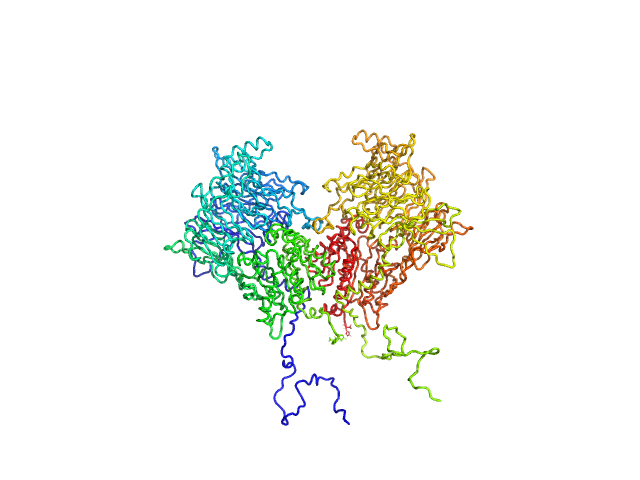
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 8 dimer, 208 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Oct 14
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
277 |
nm3 |
|
|
|
|
|
|
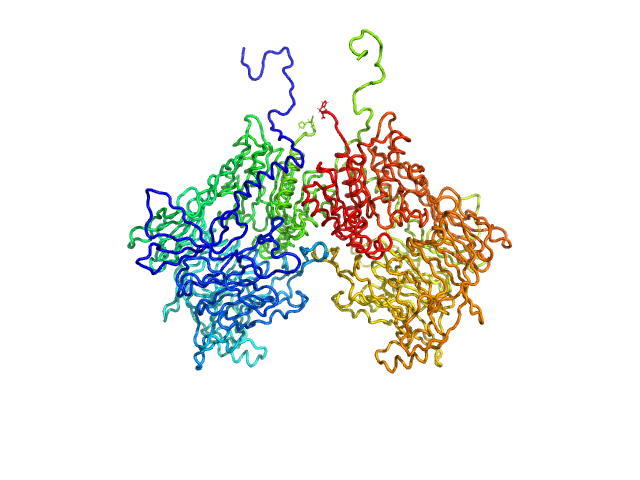
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.2 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
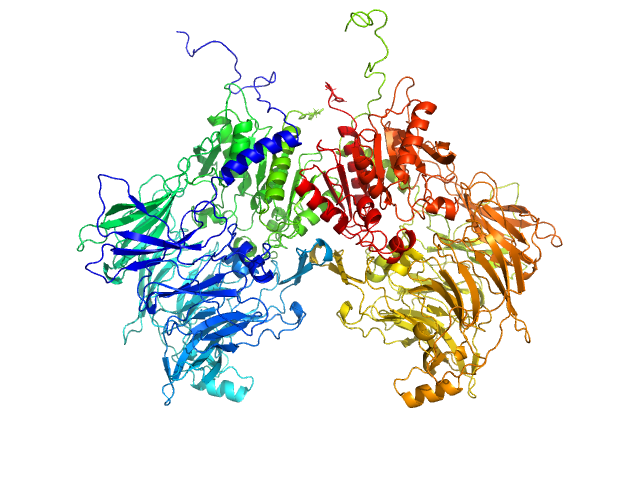
|
| Sample: |
Isoform 1 of Dipeptidyl peptidase 9 dimer, 198 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Computational study of DPP8 and DPP9: fundamental insights and inhibitor design
University of Antwerp PhD thesis c:irua:209673 (2024)
Olivier Beyens, Yann Sterckx
|
| RgGuinier |
4.1 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Ras-related protein Rab-1A monomer, 24 kDa Homo sapiens protein
|
| Buffer: |
300 mM NaCl, 2 mM 2-mercaptoethanol and 30 mM Tris-HCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2015 Nov 17
|
The SAXS Analysis of the human GTPase Rab1a
Wenhua Zhang
|
| RgGuinier |
2.4 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 19
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
6.8 |
nm |
| Dmax |
30.2 |
nm |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Pre-let-7-a-1@1 monomer, 9 kDa synthetic construct RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 14
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Pre-let-7-a-1@1 monomer, 9 kDa synthetic construct RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jun 4
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.7 |
nm |
| Dmax |
23.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
E3 ubiquitin/ISG15 ligase TRIM25 dimer, 100 kDa Homo sapiens protein
Lnczc3h7a_304-326 monomer, 8 kDa Homo sapiens RNA
|
| Buffer: |
20 mM MES, 75 mM NaCl, 1 mM TCEP, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 12
|
The molecular dissection of TRIM25's RNA-binding mechanism provides key insights into its antiviral activity.
Nat Commun 15(1):8485 (2024)
Álvarez L, Haubrich K, Iselin L, Gillioz L, Ruscica V, Lapouge K, Augsten S, Huppertz I, Choudhury NR, Simon B, Masiewicz P, Lethier M, Cusack S, Rittinger K, Gabel F, Leitner A, Michlewski G, Hentze MW, Allain FHT, Castello A, Hennig J
|
| RgGuinier |
5.8 |
nm |
| Dmax |
16.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
Tyrosine-protein kinase SYK monomer, 30 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 1 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 Nov 13
|
The mechanism of allosteric activation of SYK kinase derived from multiple phospho-ITAM-bound structures
Structure (2024)
Bradshaw W, Harris G, Gileadi O, Katis V
|
| RgGuinier |
2.2 |
nm |
| Dmax |
6.9 |
nm |
| VolumePorod |
49 |
nm3 |
|
|