|
|
|
|
|
| Sample: |
Ox40 3 UTR ADEshort monomer, 7 kDa Mus musculus RNA
|
| Buffer: |
50 mM KCl, 25 mM sodium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 7
|
NMR-derived secondary structure of the full-length Ox40 mRNA 3'UTR and its multivalent binding to the immunoregulatory RBP Roquin.
Nucleic Acids Res (2022)
Tants JN, Becker LM, McNicoll F, Müller-McNicoll M, Schlundt A
|
| RgGuinier |
1.7 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
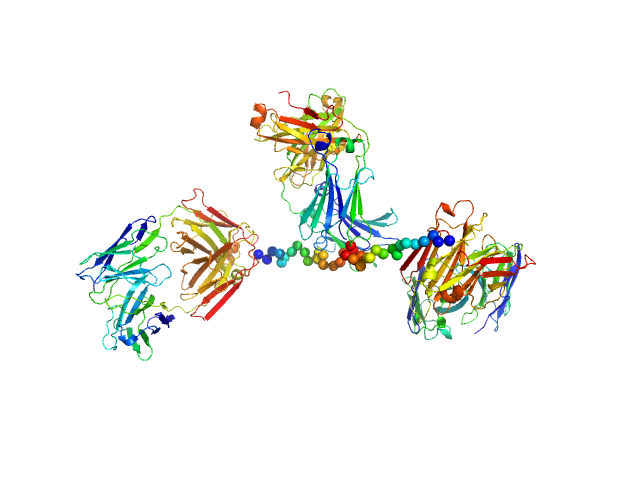
| Sample: |
Anti-prion protein monoclonal IgG2a 6D11 monomer, 145 kDa Mus musculus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies.
Sci Adv 7(48):eabj1826 (2021)
Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thapa S, Gilch S, Schätzl H, Harris DA, Triller A, Mikhaylova M, Aguzzi A, Altmeppen HC, Glatzel M
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
330 |
nm3 |
|
|
|
|
|
|
|
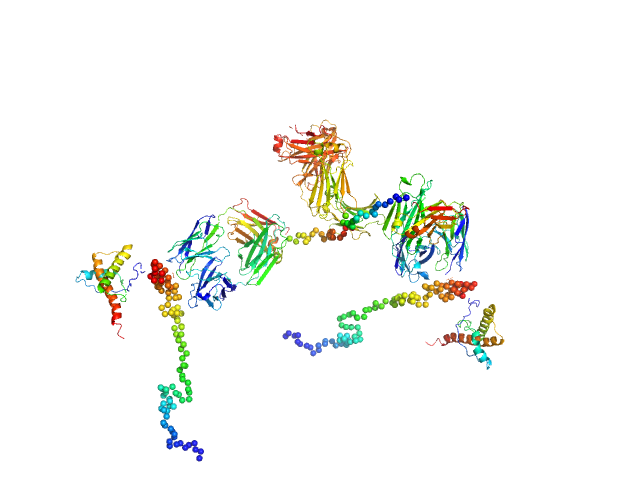
| Sample: |
Major prion protein monomer, 23 kDa Mus musculus protein
Anti-prion protein monoclonal IgG2a 6D11 monomer, 145 kDa Mus musculus protein
|
| Buffer: |
phosphate buffered saline, pH: 7.4 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 15
|
Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies.
Sci Adv 7(48):eabj1826 (2021)
Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, Schweizer M, Svergun D, Amin L, Mazzola F, Varani L, Thapa S, Gilch S, Schätzl H, Harris DA, Triller A, Mikhaylova M, Aguzzi A, Altmeppen HC, Glatzel M
|
| RgGuinier |
8.1 |
nm |
| Dmax |
24.8 |
nm |
| VolumePorod |
710 |
nm3 |
|
|
|
|
|
|
|
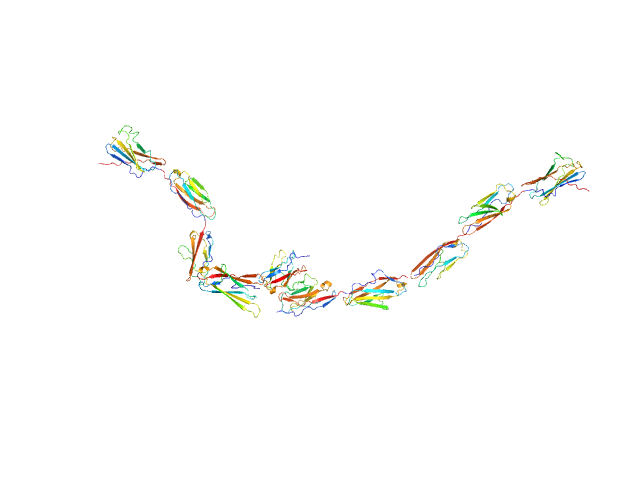
| Sample: |
Kin of IRRE-like protein 2 dimer, 106 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 3
|
Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep 37(5):109940 (2021)
Wang J, Vaddadi N, Pak JS, Park Y, Quilez S, Roman CA, Dumontier E, Thornton JW, Cloutier JF, Özkan E
|
| RgGuinier |
8.9 |
nm |
| Dmax |
39.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Kin of IRRE-like protein 3 dimer, 106 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 3
|
Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep 37(5):109940 (2021)
Wang J, Vaddadi N, Pak JS, Park Y, Quilez S, Roman CA, Dumontier E, Thornton JW, Cloutier JF, Özkan E
|
| RgGuinier |
9.3 |
nm |
| Dmax |
34.5 |
nm |
|
|
|
|
|
|
|
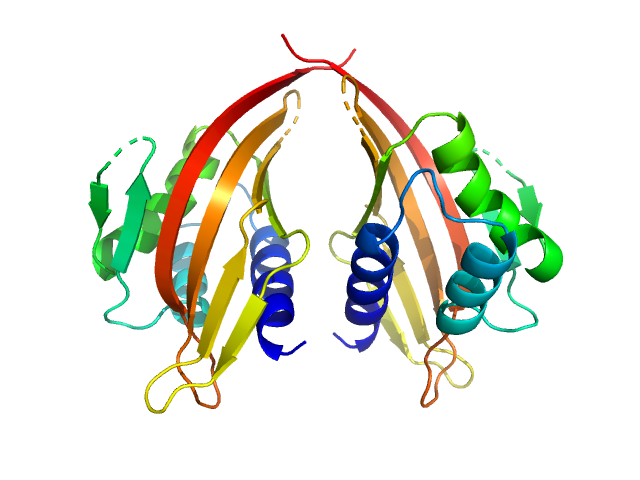
| Sample: |
Kin of IRRE-like protein 3 (Q128A) monomer, 53 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES pH 7.2, 150 mM NaCl, pH: 7.2 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Aug 3
|
Molecular and structural basis of olfactory sensory neuron axon coalescence by Kirrel receptors.
Cell Rep 37(5):109940 (2021)
Wang J, Vaddadi N, Pak JS, Park Y, Quilez S, Roman CA, Dumontier E, Thornton JW, Cloutier JF, Özkan E
|
| RgGuinier |
5.4 |
nm |
| Dmax |
21.3 |
nm |
|
|
|
|
|
|
|
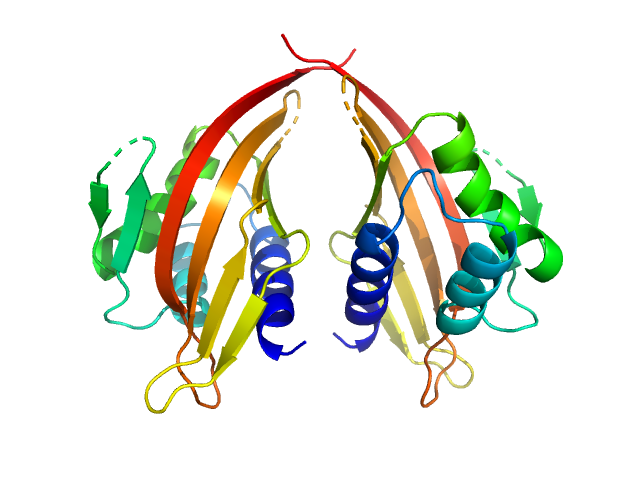
| Sample: |
LIM domain-binding protein 1 dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
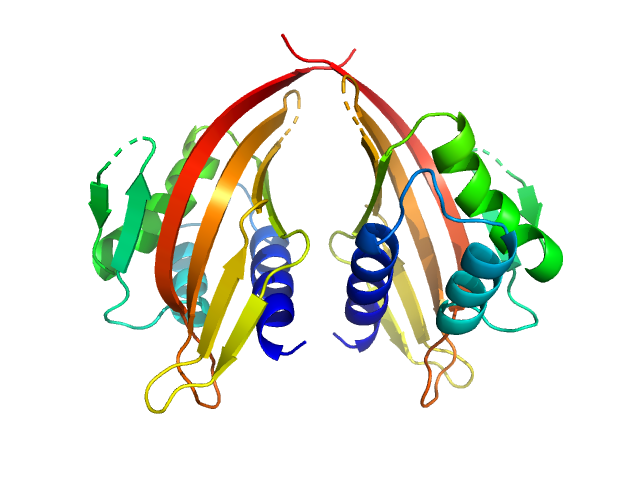
| Sample: |
LIM domain-binding protein 1, L87E dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
|
|
|
|
|
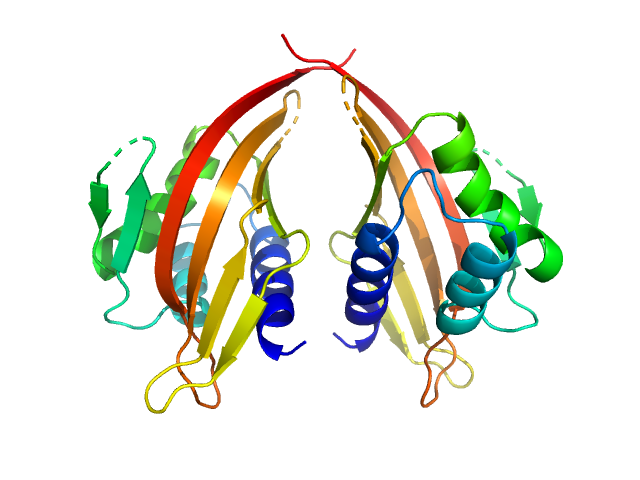
| Sample: |
LIM domain-binding protein 1, L87K dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
78 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
LIM domain-binding protein 1, L87Q dimer, 45 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM TCEP,, pH: 8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2015 Nov 19
|
Self association of LIM domain binding protein 1, LDB1
Jacqueline Matthews
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
82 |
nm3 |
|
|