|
|
|
|
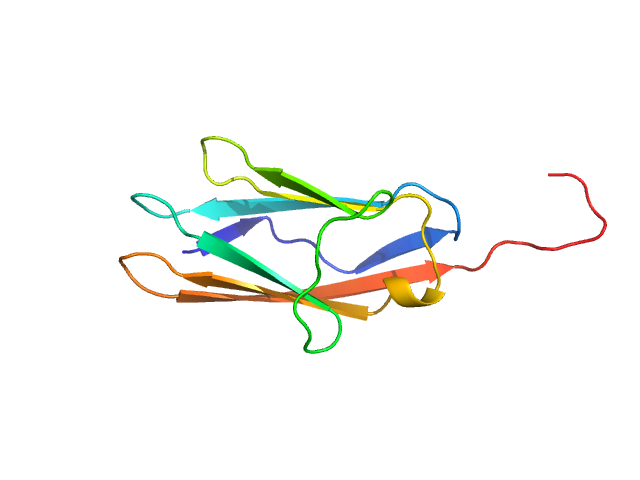
|
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
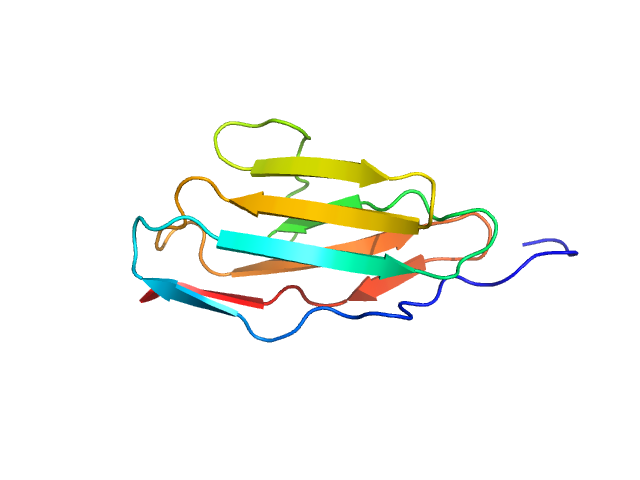
|
| Sample: |
Palladin monomer, 12 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Palladin monomer, 27 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 mM DTT, 100 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2023 Aug 29
|
Integrated structural model of the palladin-actin complex using XL-MS, docking, NMR, and SAXS.
Protein Sci 34(5):e70122 (2025)
Sargent R, Liu DH, Yadav R, Glennenmeier D, Bradford C, Urbina N, Beck MR
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 monomer, 245 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, pH: 8.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 8
|
Structural basis for regulation of CELSR1 by a compact module in its extracellular region.
Nat Commun 16(1):3972 (2025)
Bandekar SJ, Garbett K, Kordon SP, Dintzner EE, Li J, Shearer T, Sando RC, Araç D
|
| RgGuinier |
6.0 |
nm |
| Dmax |
22.5 |
nm |
| VolumePorod |
473 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cadherin EGF LAG seven-pass G-type receptor 1 dimer, 491 kDa Mus musculus protein
|
| Buffer: |
10 mM Tris, 150 mM NaCl, 1 mM CaCl2, pH: 8.5 |
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Dec 8
|
Structural basis for regulation of CELSR1 by a compact module in its extracellular region.
Nat Commun 16(1):3972 (2025)
Bandekar SJ, Garbett K, Kordon SP, Dintzner EE, Li J, Shearer T, Sando RC, Araç D
|
| RgGuinier |
16.2 |
nm |
| Dmax |
67.5 |
nm |
| VolumePorod |
722 |
nm3 |
|
|
|
|
|
|
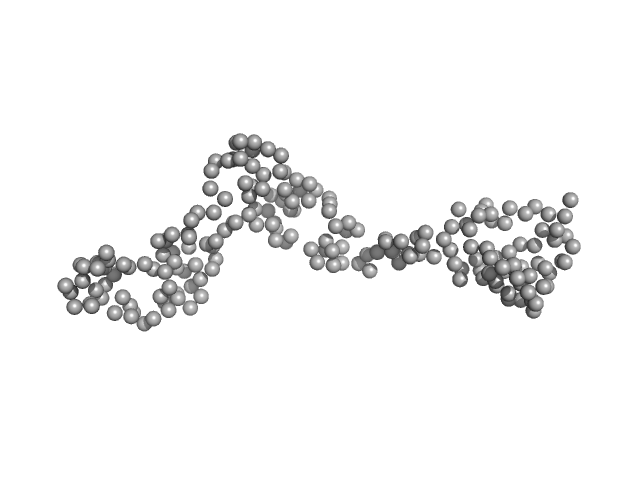
|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 21 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 42 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 40 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
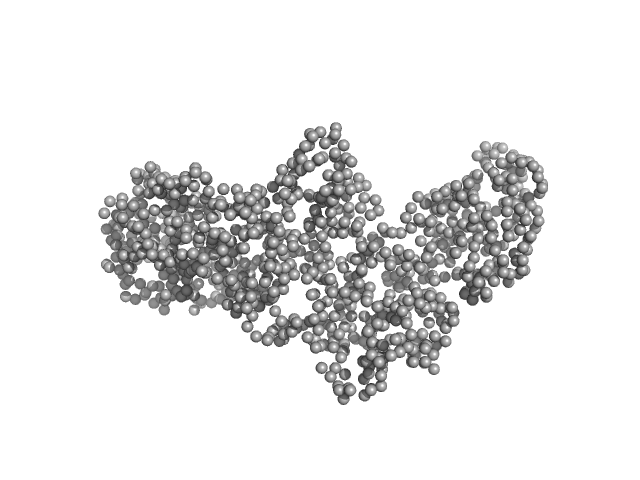
|
| Sample: |
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Oct 23
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
4.7 |
nm |
| Dmax |
17.1 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
25-mer dsRNA monomer, 16 kDa RNA
Interleukin enhancer-binding factor 2 monomer, 44 kDa Homo sapiens protein
Interleukin enhancer-binding factor 3 monomer, 66 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2023 May 9
|
Integrative structural analysis of NF45-NF90 heterodimers reveals architectural rearrangements and oligomerization on binding dsRNA.
Nucleic Acids Res 53(6) (2025)
Winterbourne S, Jayachandran U, Zou J, Rappsilber J, Granneman S, Cook AG
|
| RgGuinier |
5.7 |
nm |
| Dmax |
19.9 |
nm |
| VolumePorod |
453 |
nm3 |
|
|