|
|
|
|
|
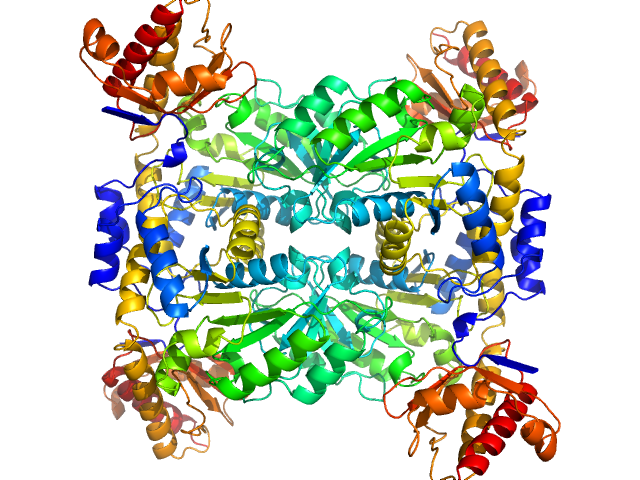
| Sample: |
L-threonine aldolase tetramer, 165 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH: 7.8 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Feb 14
|
One substrate many enzymes virtual screening uncovers missing genes of carnitine biosynthesis in human and mouse.
Nat Commun 15(1):3199 (2024)
Malatesta M, Fornasier E, Di Salvo ML, Tramonti A, Zangelmi E, Peracchi A, Secchi A, Polverini E, Giachin G, Battistutta R, Contestabile R, Percudani R
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
209 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cadherin-2 dimer, 123 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 3 mM CaCl2, 0.02% NaN3, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 1
|
Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell 187(5):1296-1311.e26 (2024)
Tsai YX, Chang NE, Reuter K, Chang HT, Yang TJ, von Bülow S, Sehrawat V, Zerrouki N, Tuffery M, Gecht M, Grothaus IL, Colombi Ciacchi L, Wang YS, Hsu MF, Khoo KH, Hummer G, Hsu SD, Hanus C, Sikora M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
496 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cadherin-2 monomer, 24 kDa Mus musculus protein
|
| Buffer: |
10 mM HEPES, 150 mM NaCl, 3 mM CaCl2, 0.02% NaN3, pH: 8 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 1
|
Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell 187(5):1296-1311.e26 (2024)
Tsai YX, Chang NE, Reuter K, Chang HT, Yang TJ, von Bülow S, Sehrawat V, Zerrouki N, Tuffery M, Gecht M, Grothaus IL, Colombi Ciacchi L, Wang YS, Hsu MF, Khoo KH, Hummer G, Hsu SD, Hanus C, Sikora M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.00, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.00, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|
|
|
|
|
|
| Sample: |
Neurofilament light polypeptide (T445N; C-terminus, amino acids 399-553), 16 kDa Mus musculus protein
|
| Buffer: |
20 mM Tris pH 8.00, 150 mM NaCl, pH: |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Sep 7
|
From isolated polyelectrolytes to star-like assemblies: the role of sequence heterogeneity on the statistical structure of the intrinsically disordered neurofilament-low tail domain.
Eur Phys J E Soft Matter 47(2):13 (2024)
Kravikass M, Koren G, Saleh OA, Beck R
|
|
|