|
|
|
|
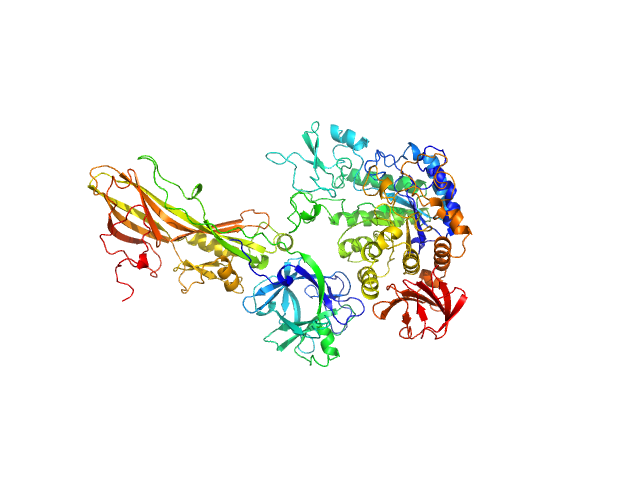
|
| Sample: |
Binary larvicide subunit BinB monomer, 53 kDa Lysinibacillus sphaericus protein
Synthetic construct (mutant, His-tagged): Mosquito-larvicidal BinAB toxin receptor protein (Neutral and basic amino acid transport protein rBAT) monomer, 65 kDa Culex quinquefasciatus protein
|
| Buffer: |
PBS buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl), pH: 7.4 |
| Experiment: |
SAXS
data collected at BL-18, INDUS-2 on 2020 Feb 24
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
4.6 |
nm |
| Dmax |
11.7 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Binary larvicide subunit BinB monomer, 53 kDa Lysinibacillus sphaericus protein
Synthetic construct (mutant, His-tagged): Mosquito-larvicidal BinAB toxin receptor protein (Neutral and basic amino acid transport protein rBAT) monomer, 65 kDa Culex quinquefasciatus protein
|
| Buffer: |
25 mM HEPES, pH 7.5, 25 mM NaCl, in 100% D2O, pH: 7.5 |
| Experiment: |
SANS
data collected at SANS-I facility, Dhruva Reactor, Bhabha Atomic Research Centre on 2019 Apr 26
|
Liposome-Based Study Provides Insight into Cellular Internalization Mechanism of Mosquito-Larvicidal BinAB Toxin.
J Membr Biol (2020)
Sharma M, Kumar A, Kumar V
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
174 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Major tail protein monomer, 40 kDa Salmonella virus Chi protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 0.03 % NaN3, 5.0 % glycerol, pH: 7.8 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Mar 8
|
The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun 11(1):3748 (2020)
Hardy JM, Dunstan RA, Grinter R, Belousoff MJ, Wang J, Pickard D, Venugopal H, Dougan G, Lithgow T, Coulibaly F
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Wildtype preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Wildtype preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C15 deletion preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
C15 deletion preQ1 riboswitch in Bacillus subtilis monomer, 11 kDa Bacillus subtilis RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
2.0 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Wildtype preQ1 riboswitch in Thermoanaerobacter tengcongensis monomer, 11 kDa Caldanaerobacter subterraneus subsp. … RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Wildtype preQ1 riboswitch in Thermoanaerobacter tengcongensis monomer, 11 kDa Caldanaerobacter subterraneus subsp. … RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.6 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
13 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Wildtype preQ1 riboswitch in Thermoanaerobacter tengcongensis monomer, 11 kDa Caldanaerobacter subterraneus subsp. … RNA
|
| Buffer: |
50 mM potassium phosphate, 2 mM MgCl2, 50 mM KCl, pH: 6.5 |
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2018 Mar 23
|
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.
J Mol Biol 432(16):4523-4543 (2020)
Gong Z, Yang S, Dong X, Yang QF, Zhu YL, Xiao Y, Tang C
|
| RgGuinier |
1.7 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
11 |
nm3 |
|
|