|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.4 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.5 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.8 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.6 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
24 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
25 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
26 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A), 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alpha/beta fold hydrolase monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM NaCl, 10 mM HEPES, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Apr 11
|
Similar but Distinct—Biochemical Characterization of the
Staphylococcus aureus
Serine Hydrolases FphH
and FphI
Proteins: Structure, Function, and Bioinformatics (2024)
Fellner M, Randall G, Bitac I, Warrender A, Sethi A, Jelinek R, Kass I
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
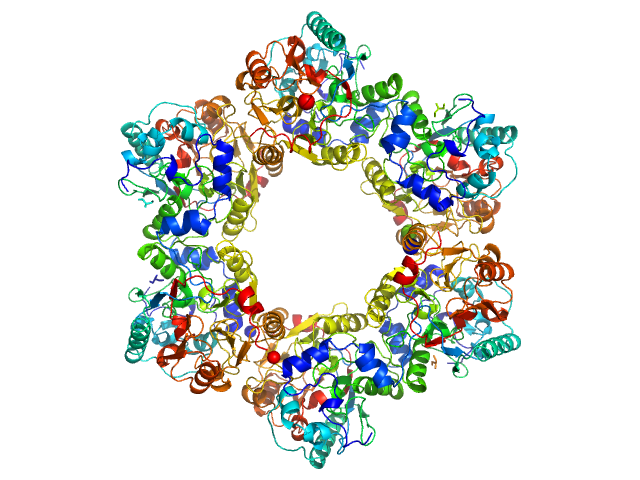
|
| Sample: |
2-nitroimidazole nitrohydrolase (T2I, G14D, K73R) hexamer, 258 kDa Mycobacterium sp. (strain … protein
|
| Buffer: |
25 mM Tris-HCl, 150 mM NaCl, 5% (v/v) glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Aug 6
|
Structural insights into the enzymatic breakdown of azomycin-derived antibiotics by 2-nitroimdazole hydrolase (NnhA).
Commun Biol 7(1):1676 (2024)
Ahmed FH, Liu JW, Royan S, Warden AC, Esquirol L, Pandey G, Newman J, Scott C, Peat TS
|
| RgGuinier |
4.6 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
363 |
nm3 |
|
|