|
|
|
|
|
| Sample: |
Zinc finger and BTB domain-containing protein 8A.1-A dimer, 35 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 4
|
Dynamic BTB-domain filaments promote clustering of ZBTB proteins.
Mol Cell 84(13):2490-2510.e9 (2024)
Mance L, Bigot N, Zhamungui Sánchez E, Coste F, Martín-González N, Zentout S, Biliškov M, Pukało Z, Mishra A, Chapuis C, Arteni AA, Lateur A, Goffinont S, Gaudon V, Talhaoui I, Casuso I, Beaufour M, Garnier N, Artzner F, Cadene M, Huet S, Castaing B, Suskiewicz MJ
|
| RgGuinier |
22.3 |
nm |
| Dmax |
99.6 |
nm |
|
|
|
|
|
|
|
| Sample: |
Zinc finger and BTB domain-containing protein 8A.1-A (S103R mutant) dimer, 35 kDa Xenopus laevis protein
|
| Buffer: |
20 mM Hepes, 150 mM NaCl, 0.5 mM TCEP, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Dec 4
|
Dynamic BTB-domain filaments promote clustering of ZBTB proteins.
Mol Cell 84(13):2490-2510.e9 (2024)
Mance L, Bigot N, Zhamungui Sánchez E, Coste F, Martín-González N, Zentout S, Biliškov M, Pukało Z, Mishra A, Chapuis C, Arteni AA, Lateur A, Goffinont S, Gaudon V, Talhaoui I, Casuso I, Beaufour M, Garnier N, Artzner F, Cadene M, Huet S, Castaing B, Suskiewicz MJ
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
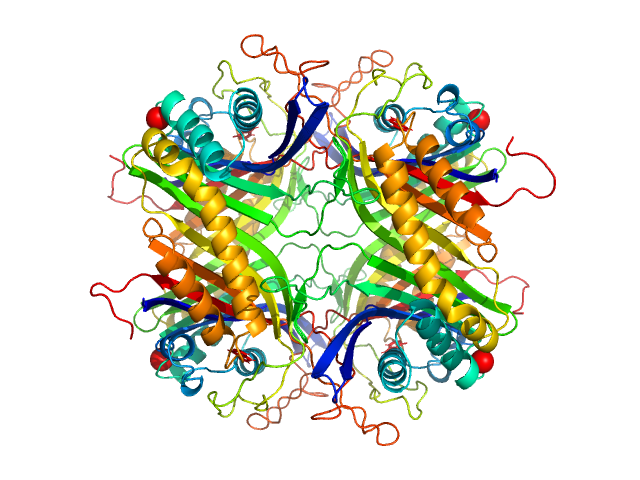
| Sample: |
Uricase tetramer, 136 kDa Aspergillus flavus protein
|
| Buffer: |
100 mM Tris, 150 mM NaCl, pH: 8 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.3 |
nm |
|
|
|
|
|
|
|
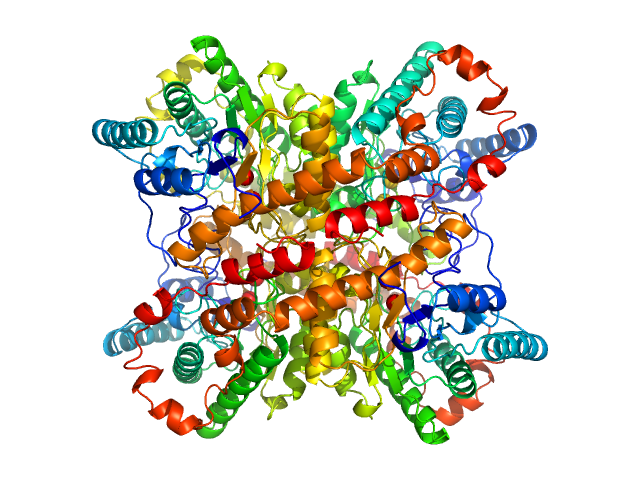
| Sample: |
Xylose isomerase tetramer, 173 kDa Streptomyces rubiginosus protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 1 mM MgCl2, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.1 |
nm |
|
|
|
|
|
|
|
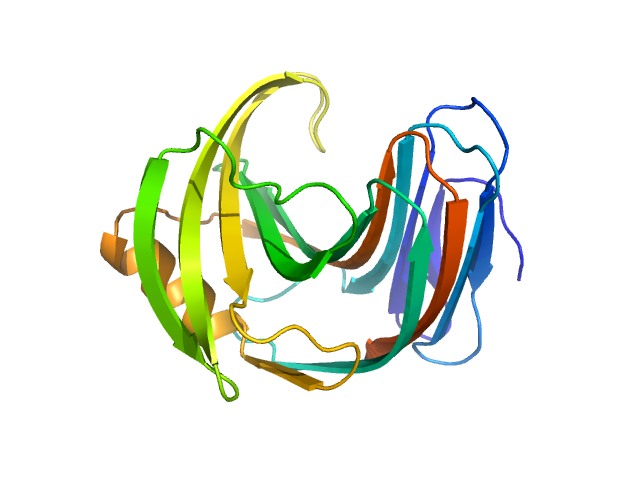
| Sample: |
Endo-1,4-beta-xylanase monomer, 21 kDa Trichoderma longibrachiatum protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, pH: 7.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.6 |
nm |
| Dmax |
5.2 |
nm |
|
|
|
|
|
|
|
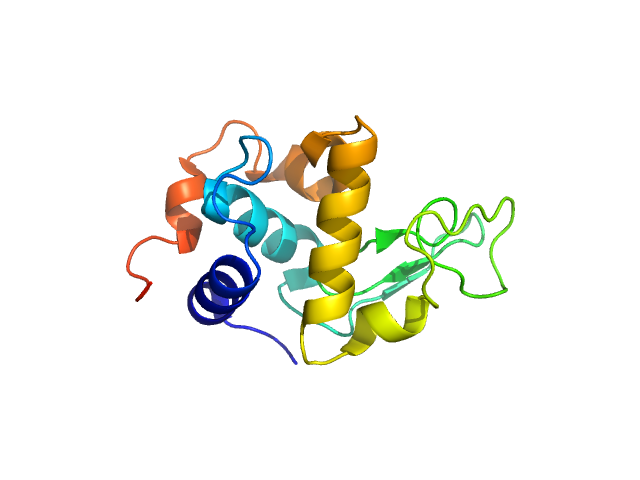
| Sample: |
Lysozyme C monomer, 14 kDa Gallus gallus protein
|
| Buffer: |
50 mM sodium citrate, 150 mM NaCl, pH: 4.5 |
| Experiment: |
SANS
data collected at (Consensus SAS), Multi-facility, Multiple countries on 2024 Feb 4
|
Benchmarking predictive methods for small-angle X-ray scattering from atomic coordinates of proteins using maximum likelihood consensus data
IUCrJ 11(5) (2024)
Trewhella J, Vachette P, Larsen A
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.8 |
nm |
|
|
|
|
|
|
|
| Sample: |
Pigeon iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
|
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|