|
|
|
|
|
| Sample: |
Stem loop 2 and 3 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
Nucleoprotein dimer, 30 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 with AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 22 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AU extension in the 5'-genomic end of SARS-CoV-2 monomer, 7 kDa Severe acute respiratory … RNA
Nucleoprotein dimer, 30 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoprotein monomer, 15 kDa Severe acute respiratory … protein
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM HEPES, 75 mM KCl, 2.5 mM NaNO3, pH: 7.2 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Nov 29
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements
Karthikeyan Dhamotharan
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|
|
|
|
|
|
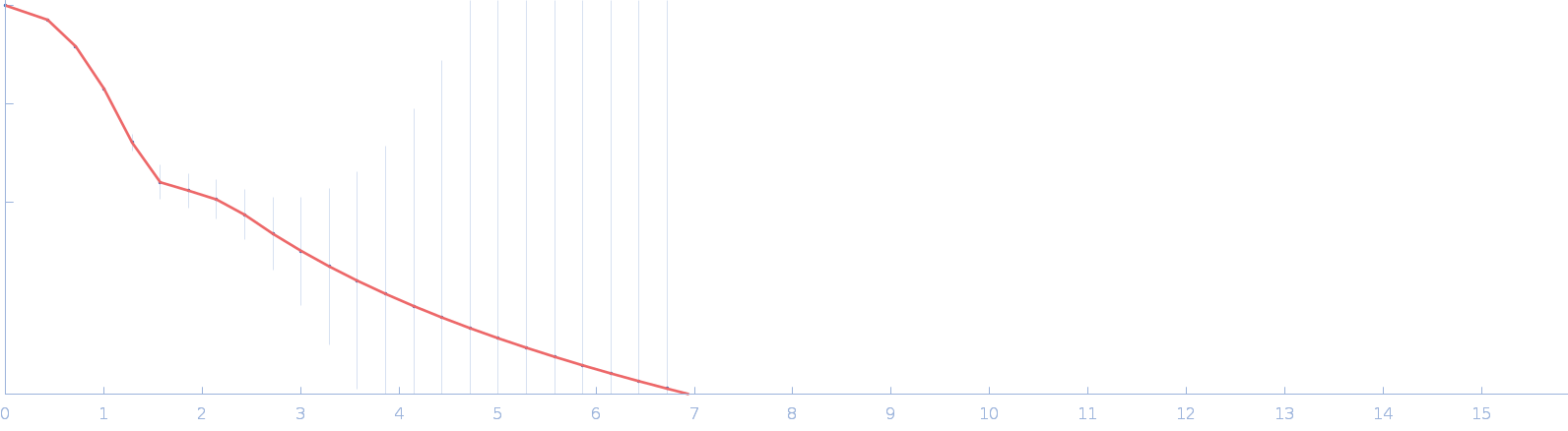
| Sample: |
DUF4374 domain-containing protein monomer, 46 kDa Bacteroides thetaiotaomicron (strain … protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2021 May 9
|
Iron acquisition by a commensal bacterium modifies host nutritional immunity during Salmonella infection.
Cell Host Microbe 31(10):1639-1654.e10 (2023)
Spiga L, Fansler RT, Perera YR, Shealy NG, Munneke MJ, David HE, Torres TP, Lemoff A, Ran X, Richardson KL, Pudlo N, Martens EC, Folta-Stogniew E, Yang ZJ, Skaar EP, Byndloss MX, Chazin WJ, Zhu W
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
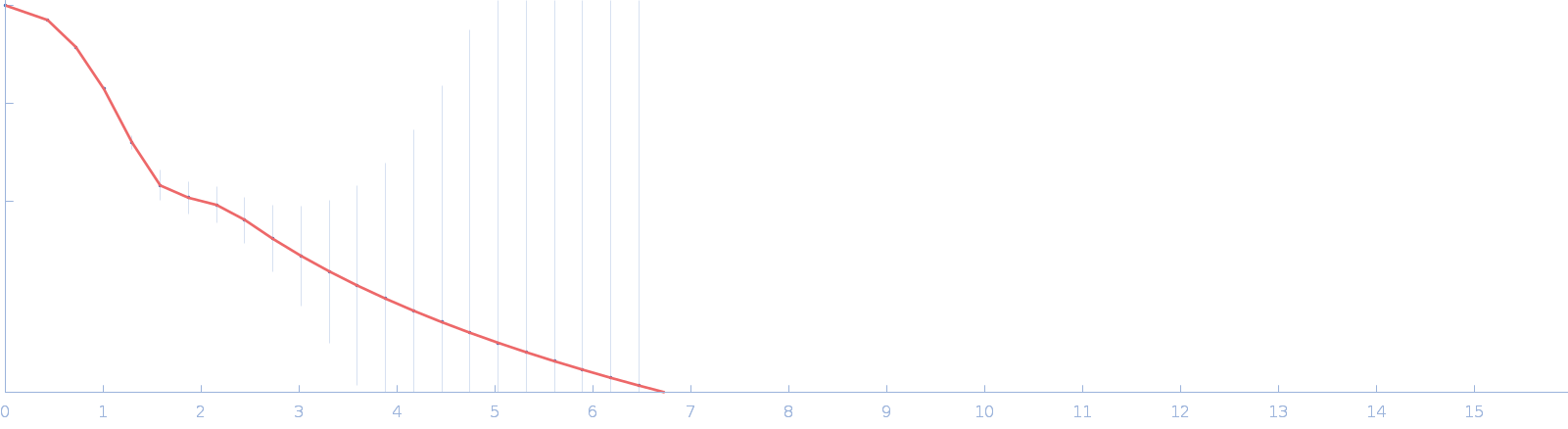
| Sample: |
DUF4374 domain-containing protein monomer, 46 kDa Bacteroides thetaiotaomicron (strain … protein
|
| Buffer: |
20 mM Tris 150 mM NaCl, pH: 8 |
| Experiment: |
SAXS
data collected at 12-ID-B, Advanced Photon Source (APS), Argonne National Laboratory on 2021 May 9
|
Iron acquisition by a commensal bacterium modifies host nutritional immunity during Salmonella infection.
Cell Host Microbe 31(10):1639-1654.e10 (2023)
Spiga L, Fansler RT, Perera YR, Shealy NG, Munneke MJ, David HE, Torres TP, Lemoff A, Ran X, Richardson KL, Pudlo N, Martens EC, Folta-Stogniew E, Yang ZJ, Skaar EP, Byndloss MX, Chazin WJ, Zhu W
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
77 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Frameshifting pseudoknot from SARS CoV2, wild type monomer, 23 kDa RNA
|
| Buffer: |
50 mM MOPS, 130 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2020 May 15
|
Atomistic structure of the SARS-CoV-2 pseudoknot in solution from SAXS-driven molecular dynamics.
Nucleic Acids Res (2023)
He W, San Emeterio J, Woodside MT, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
40 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Variant: non frameshifting pseudoknot from SARS CoV2 genome monomer, 23 kDa RNA
|
| Buffer: |
50 mM MOPS, 130 mM KCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at 16-ID (LiX), National Synchrotron Light Source II (NSLS-II) on 2020 Jun 12
|
Atomistic structure of the SARS-CoV-2 pseudoknot in solution from SAXS-driven molecular dynamics.
Nucleic Acids Res (2023)
He W, San Emeterio J, Woodside MT, Kirmizialtin S, Pollack L
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.5 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
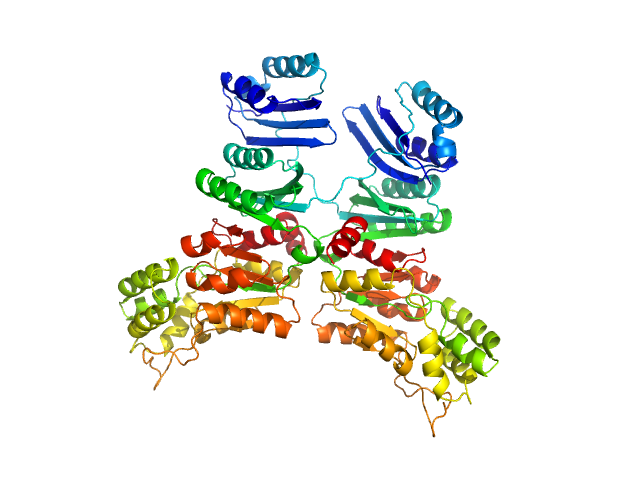
| Sample: |
Phosphoserine phosphatase dimer, 88 kDa Mycobacterium marinum (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 6
|
A morpheein equilibrium regulates catalysis in phosphoserine phosphatase SerB2 from Mycobacterium tuberculosis.
Commun Biol 6(1):1024 (2023)
Pierson E, De Pol F, Fillet M, Wouters J
|
| RgGuinier |
3.2 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoserine phosphatase (G31R, G152E) dimer, 88 kDa Mycobacterium avium (strain … protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 1 mM TCEP, pH: 7.4 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 6
|
A morpheein equilibrium regulates catalysis in phosphoserine phosphatase SerB2 from Mycobacterium tuberculosis.
Commun Biol 6(1):1024 (2023)
Pierson E, De Pol F, Fillet M, Wouters J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
138 |
nm3 |
|
|