|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 26 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Jul 19
|
Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res (2023)
Hollmann NM, Jagtap PKA, Linse JB, Ullmann P, Payr M, Murciano B, Simon B, Hub JS, Hennig J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 63 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2019 Sep 30
|
Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res (2023)
Hollmann NM, Jagtap PKA, Linse JB, Ullmann P, Payr M, Murciano B, Simon B, Hub JS, Hennig J
|
| RgGuinier |
4.9 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
136 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 57 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Sep 30
|
Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res (2023)
Hollmann NM, Jagtap PKA, Linse JB, Ullmann P, Payr M, Murciano B, Simon B, Hub JS, Hennig J
|
| RgGuinier |
4.4 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
160 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Upstream of N-ras, isoform A monomer, 26 kDa Drosophila melanogaster protein
PolyA-15mer monomer, 5 kDa RNA
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 8
|
Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res (2023)
Hollmann NM, Jagtap PKA, Linse JB, Ullmann P, Payr M, Murciano B, Simon B, Hub JS, Hennig J
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
37 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA (Zinc finger protein 410 recognition sequence) monomer, 11 kDa DNA
|
| Buffer: |
20 mM Tris, 250 mM NaCl, 0.1% v/v β-mercaptoethanol, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2020 Sep 30
|
Allosteric autoregulation of DNA binding via a DNA-mimicking protein domain: a biophysical study of ZNF410-DNA interaction using small angle X-ray scattering.
Nucleic Acids Res (2023)
Kaur G, Ren R, Hammel M, Horton JR, Yang J, Cao Y, He C, Lan F, Lan X, Blobel GA, Blumenthal RM, Zhang X, Cheng X
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.8 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|

|
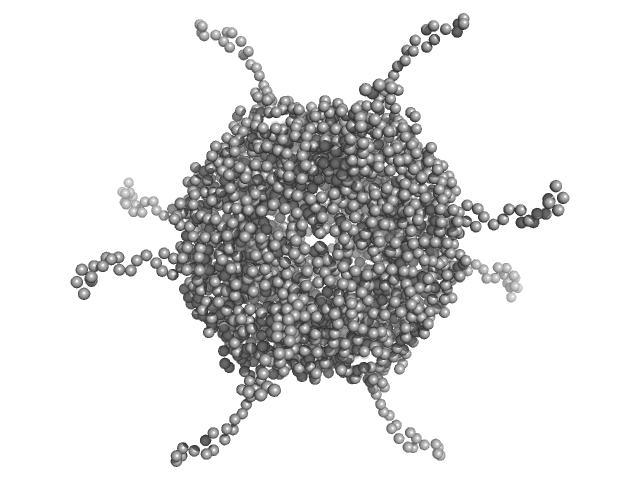
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.5 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
454 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.4 |
nm |
| Dmax |
18.4 |
nm |
| VolumePorod |
440 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
464 |
nm3 |
|
|
|
|
|
|

|
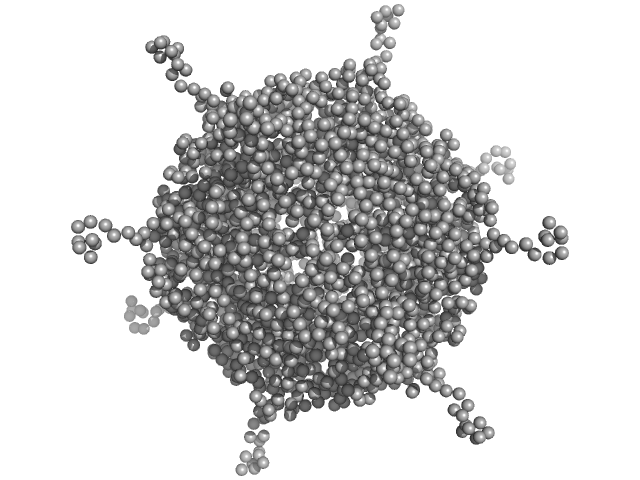
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
439 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA protection during starvation, DPS (Ferritin superfamily) dodecamer, 270 kDa Deinococcus grandis protein
|
| Buffer: |
50 mM MOPS-NaOH, 230 mM NaCl, pH: 7 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 8
|
Controlled modulation of the dynamics of the Deinococcus grandis Dps N-terminal tails by divalent metals.
Protein Sci :e4567 (2023)
Guerra JPL, Blanchet CE, Vieira BJC, Waerenborgh JC, Jones NC, Hoffmann SV, Pereira AS, Tavares P
|
| RgGuinier |
4.2 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
427 |
nm3 |
|
|