|
|
|
|
|
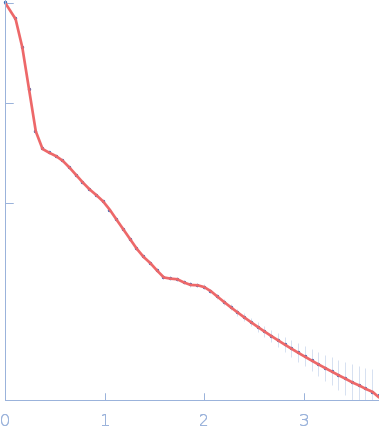
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jul 15
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
10.3 |
nm |
| Dmax |
31.0 |
nm |
| VolumePorod |
1170 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Δ212-217) octamer, 526 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jun 25
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
13.7 |
nm |
| Dmax |
25.5 |
nm |
| VolumePorod |
961 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (Δ212-217) octamer, 528 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jun 26
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
10.8 |
nm |
| Dmax |
30.3 |
nm |
| VolumePorod |
1260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jul 2
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
9.5 |
nm |
| Dmax |
28.3 |
nm |
| VolumePorod |
1010 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] hemoprotein beta-component tetramer, 256 kDa Escherichia coli (strain … protein
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jun 27
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
8.2 |
nm |
| Dmax |
25.1 |
nm |
| VolumePorod |
839 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component octamer, 530 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jul 15
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
7.9 |
nm |
| Dmax |
27.6 |
nm |
| VolumePorod |
1060 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (E121C, C162T, C552S, N556C) octamer, 566 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jul 15
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
8.0 |
nm |
| Dmax |
25.3 |
nm |
| VolumePorod |
827 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Sulfite reductase [NADPH] flavoprotein alpha-component (Δ212-217) octamer, 526 kDa Escherichia coli (strain … protein
|
| Buffer: |
50 mM potassium phosphate, 100 mM NaCl, 1 mM EDTA, pH: 7.8
|
| Experiment: |
SANS
data collected at EQ-SANS, Spallation Neutron Source on 2022 Jul 15
|
Domain crossover in the reductase subunit of NADPH-dependent assimilatory sulfite reductase.
J Struct Biol 215(4):108028 (2023)
Walia N, Murray DT, Garg Y, He H, Weiss KL, Nagy G, Elizabeth Stroupe M
|
| RgGuinier |
7.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
952 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab, H3426A (3C-like proteinase nsp5 - H163A mutant) dimer, 67 kDa Severe acute respiratory … protein
|
| Buffer: |
25 mM HEPES, 1 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Mar 24
|
The H163A mutation unravels an oxidized conformation of the SARS-CoV-2 main protease.
Nat Commun 14(1):5625 (2023)
Tran N, Dasari S, Barwell SAE, McLeod MJ, Kalyaanamoorthy S, Holyoak T, Ganesan A
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.2 |
nm |
| VolumePorod |
74 |
nm3 |
|
|