|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase dimer, 114 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase dimer, 114 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, 1 mM nicotinamide, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase dimer, 114 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, 1 mM nicotinic acid, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase dimer, 114 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, 1 mM phosphoribosyl pyrophosphate, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.9 |
nm |
| VolumePorod |
173 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase Δ42-51 dimer, 111 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
159 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase Δ42-51 dimer, 111 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, 1 mM nicotinamide, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.9 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase Δ42-51 dimer, 111 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, 1 mM nicotinic acid, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.2 |
nm |
| VolumePorod |
152 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nicotinamide phosphoribosyltransferase Δ42-51 dimer, 111 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 500 mM NaCl, 6 mM MgCl2, 5% (v/v) glycerol, 1 mM phosphoribosyl pyrophosphate, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 May 20
|
Identification of structural determinants of nicotinamide phosphoribosyl transferase (NAMPT) activity and substrate selectivity.
J Struct Biol :108004 (2023)
Houry D, Raasakka A, Ferrario E, Niere M, Bifulco E, Kursula P, Ziegler M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|
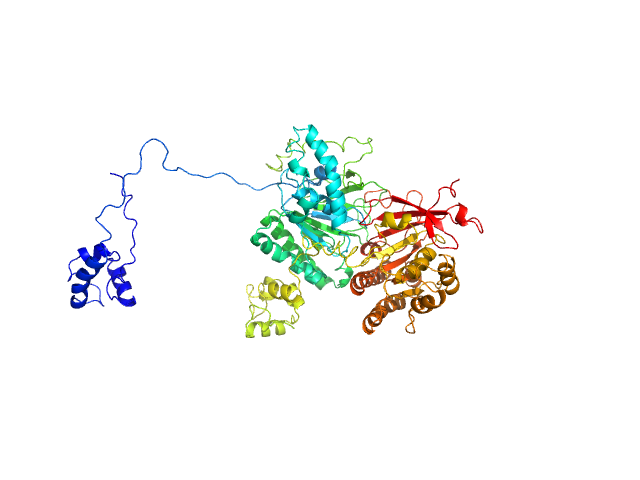
|
| Sample: |
Isoform 1 of DNA repair protein RAD51 homolog 3 monomer, 40 kDa Homo sapiens protein
DNA repair protein XRCC3 monomer, 38 kDa Homo sapiens protein
|
| Buffer: |
10 mM HEPES pH 8, 100 mM NaCl, 2.5 mM ATP, 2.5 mM MgCl2, and 0.1 mM Na3VO4, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 May 5
|
RAD51C-XRCC3 structure and cancer patient mutations define DNA replication roles
Nature Communications 14(1) (2023)
Longo M, Roy S, Chen Y, Tomaszowski K, Arvai A, Pepper J, Boisvert R, Kunnimalaiyaan S, Keshvani C, Schild D, Bacolla A, Williams G, Tainer J, Schlacher K
|
| RgGuinier |
3.6 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
132 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial monomer, 70 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 1
|
Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22
Jahangir Alam
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
160 |
nm3 |
|
|