|
|
|
|

|
| Sample: |
dimeric Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22, mitochondrial dimer, 141 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
40 mM Tris pH 7.5, 500 mM NaCl, 5% glycerol, 2.5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 May 1
|
Probable S-adenosyl-L-methionine-dependent RNA methyltransferase RSM22
Jahangir Alam
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.2 |
nm |
| VolumePorod |
516 |
nm3 |
|
|
|
|
|
|
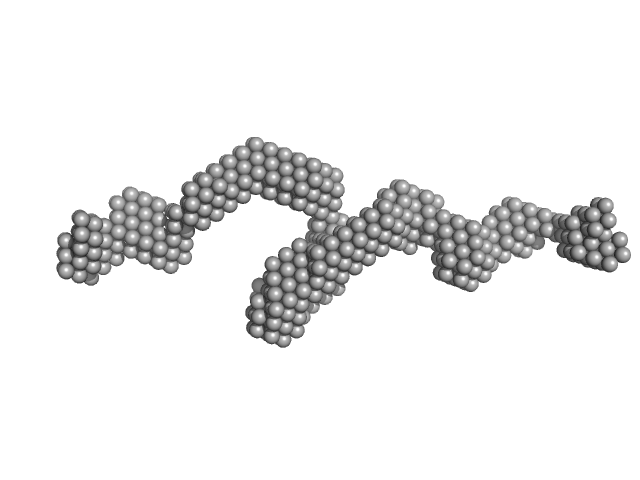
|
| Sample: |
Sperm-associated antigen 1 monomer, 104 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
7.3 |
nm |
| Dmax |
34.0 |
nm |
| VolumePorod |
374 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RuvB-like 1 hexamer, 311 kDa Homo sapiens protein
RuvB-like 2 hexamer, 311 kDa Homo sapiens protein
Sperm-associated antigen 1 monomer, 104 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
6.8 |
nm |
| Dmax |
34.8 |
nm |
| VolumePorod |
1800 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
RuvB-like 1 hexamer, 311 kDa Homo sapiens protein
RuvB-like 2 hexamer, 311 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 3
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
6.3 |
nm |
| Dmax |
23.4 |
nm |
| VolumePorod |
1550 |
nm3 |
|
|
|
|
|
|
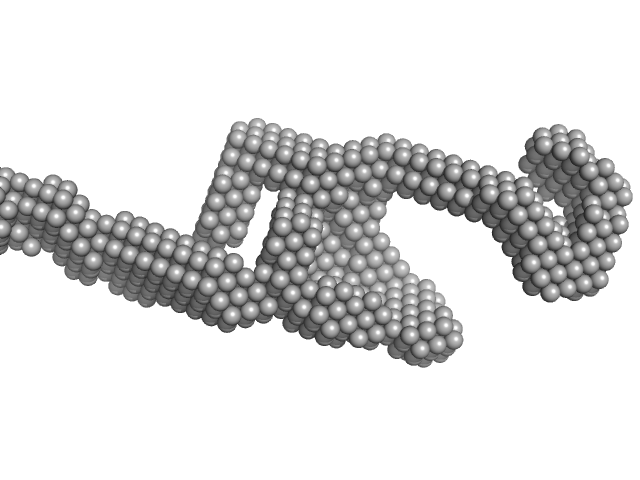
|
| Sample: |
Sperm-associated antigen 1 monomer, 104 kDa Homo sapiens protein
PIH1 domain-containing protein 2 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
7.0 |
nm |
| Dmax |
30.9 |
nm |
| VolumePorod |
664 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
PIH1 domain-containing protein 2 monomer, 36 kDa Homo sapiens protein
Sperm-associated antigen 1 monomer, 35 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 1% glycerol, 5 mM TCEP, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2019 Mar 21
|
R2SP
Raphael Dos Santos Morais
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.3 |
nm |
| VolumePorod |
123 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.8 |
nm |
| VolumePorod |
181 |
nm3 |
|
|
|
|
|
|
|
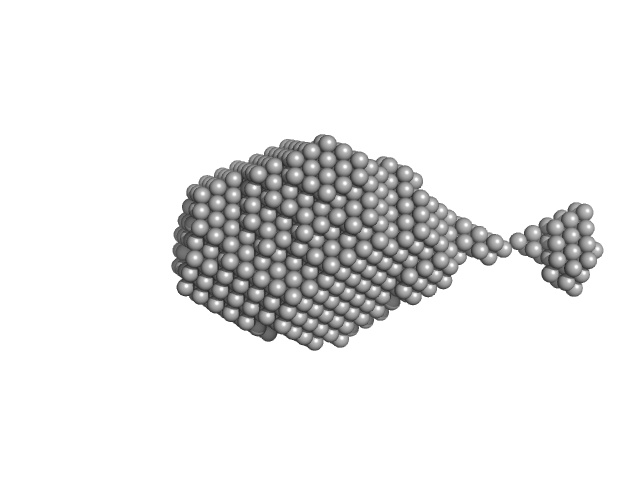
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
|
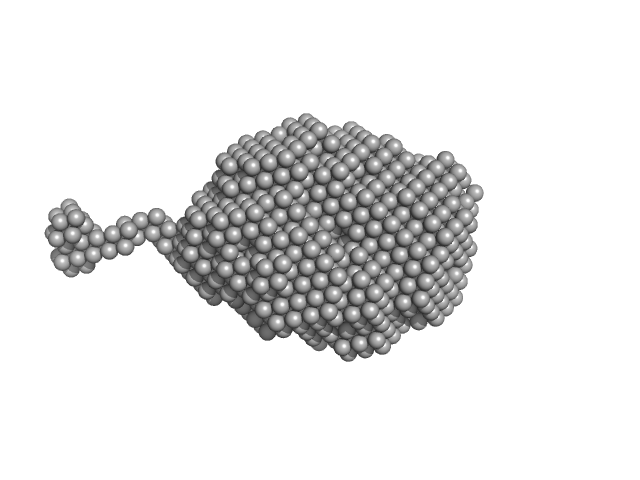
| Sample: |
Aromatic-L-amino-acid decarboxylase (M17V) dimer, 108 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, pH: 7.4
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2022 Feb 28
|
Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: implication in AADC
deficiency
Protein Science (2023)
Bisello G, Ribeiro R, Perduca M, Belviso B, Polverino de' Laureto P, Giorgetti A, Caliandro R, Bertoldi M
|
| RgGuinier |
3.0 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
154 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
6S RNA (SsrS gene) monomer, 59 kDa Escherichia coli RNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM KCl, 5 mM MgCl2, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 4
|
Probing the conformational changes of in vivo overexpressed cell cycle regulator 6S ncRNA
Frontiers in Molecular Biosciences 10 (2023)
Makraki E, Miliara S, Pagkalos M, Kokkinidis M, Mylonas E, Fadouloglou V
|
| RgGuinier |
6.1 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
175 |
nm3 |
|
|