|
|
|
|
|
| Sample: |
6S RNA (SsrS gene) monomer, 59 kDa Escherichia coli RNA
product RNA from E. coli 6S monomer, 7 kDa Escherichia coli RNA
|
| Buffer: |
20 mM Tris-HCl, 200 mM KCl, 5 mM MgCl2, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Apr 4
|
Probing the conformational changes of in vivo overexpressed cell cycle regulator 6S ncRNA
Frontiers in Molecular Biosciences 10 (2023)
Makraki E, Miliara S, Pagkalos M, Kokkinidis M, Mylonas E, Fadouloglou V
|
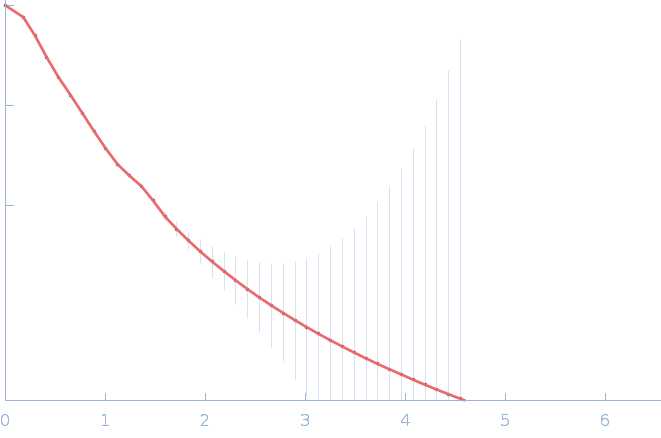
| RgGuinier |
4.9 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
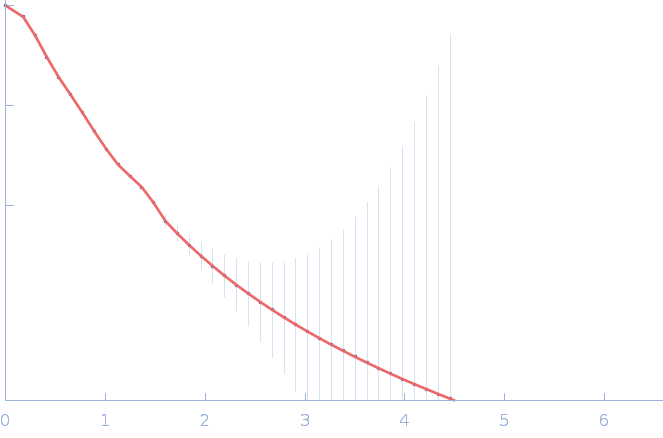
| RgGuinier |
4.9 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
233 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
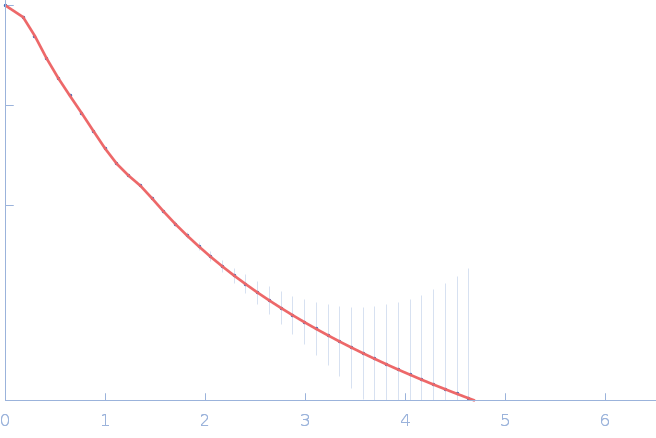
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
225 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
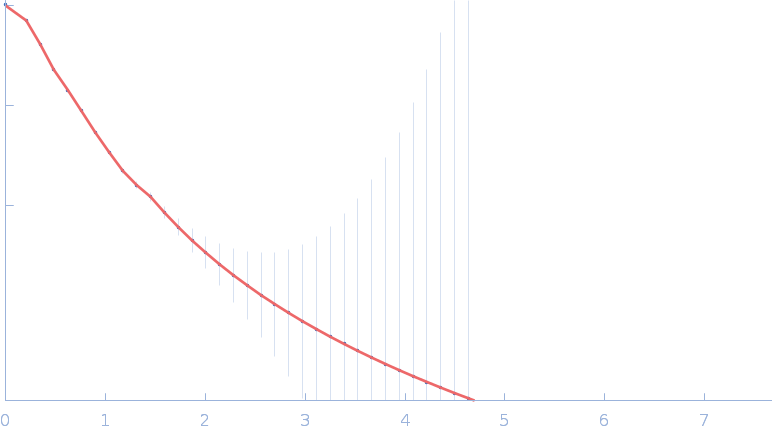
| RgGuinier |
4.9 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
243 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nitrogen fixation regulatory protein (Q409L) dimer, 115 kDa Azotobacter vinelandii protein
|
| Buffer: |
50 mM Bis-Tris, 100 mM (NH4)2SO4, 10% glycerol, 5 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2022 Jul 5
|
Structural insights into redox signal transduction mechanisms in the control of nitrogen fixation by the NifLA system
Proceedings of the National Academy of Sciences 120(30) (2023)
Boyer N, Tokmina-Lukaszewska M, Bueno Batista M, Mus F, Dixon R, Bothner B, Peters J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
224 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA-binding protein 5 (I107T, C191G) monomer, 14 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 400 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Sep 26
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
1.7 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
28 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
RNA Binding Motif protein 5 (I107T, C191G) monomer, 26 kDa Homo sapiens protein
|
| Buffer: |
20 mM MES, 400 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2017 Feb 9
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
|
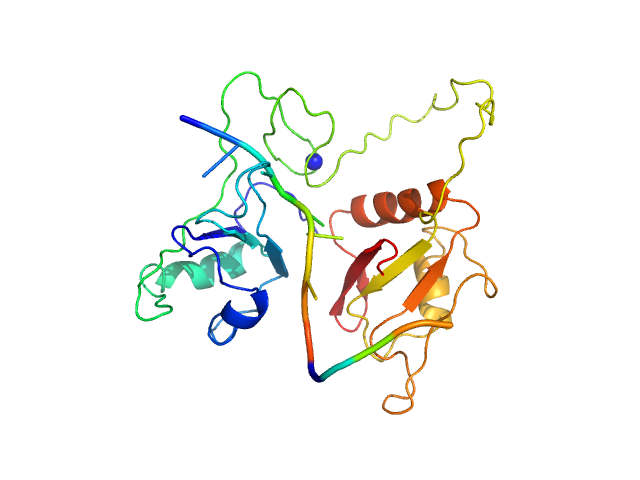
| Sample: |
RNA Binding Motif protein 5 (I107T, C191G) monomer, 26 kDa Homo sapiens protein
Caspase-2 derived RNA GGCU_12 monomer, 4 kDa RNA
|
| Buffer: |
20 mM MES, 100 mM NaCl, 1 mM DTT, pH: 6.5
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, SFB 1035, Technische Universität München on 2017 Mar 1
|
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.
Nat Commun 14(1):4233 (2023)
Soni K, Jagtap PKA, Martínez-Lumbreras S, Bonnal S, Geerlof A, Stehle R, Simon B, Valcárcel J, Sattler M
|
| RgGuinier |
2.2 |
nm |
| Dmax |
7.5 |
nm |
| VolumePorod |
42 |
nm3 |
|
|
|
|
|
|
|
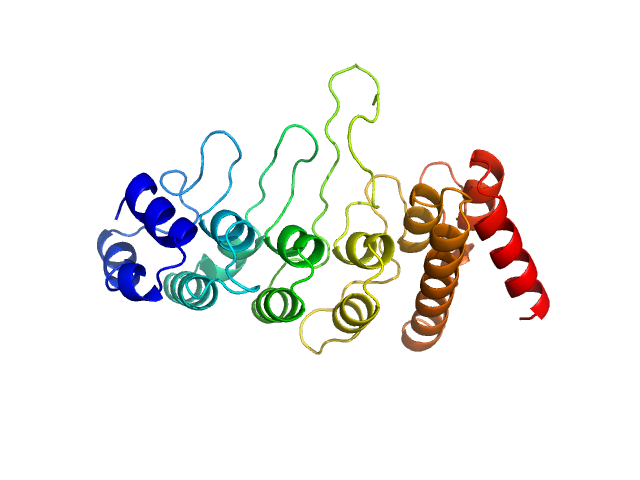
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 28 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transient receptor potential cation channel subfamily V member 4 monomer, 43 kDa Gallus gallus protein
|
| Buffer: |
20 mM Tris, 300 mM NaCl, 10 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Dec 16
|
Crosstalk between regulatory elements in disordered TRPV4 N-terminus modulates lipid-dependent channel activity
Nature Communications 14(1) (2023)
Goretzki B, Wiedemann C, McCray B, Schäfer S, Jansen J, Tebbe F, Mitrovic S, Nöth J, Cabezudo A, Donohue J, Jeffries C, Steinchen W, Stengel F, Sumner C, Hummer G, Hellmich U
|
| RgGuinier |
3.7 |
nm |
| Dmax |
19.0 |
nm |
| VolumePorod |
59 |
nm3 |
|
|