|
|
|
|
|
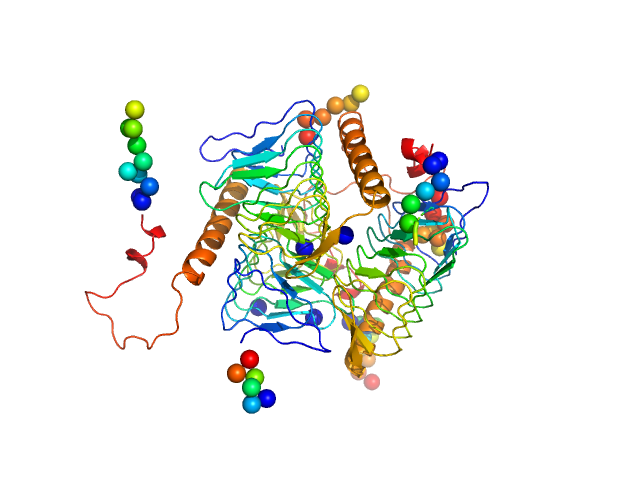
| Sample: |
Bacterial transferase hexapeptide repeat protein trimer, 66 kDa Acinetobacter baumannii (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2021 Jan 29
|
Mechanistic and structural insights into the bifunctional enzyme PaaY from Acinetobacter baumannii.
Structure (2023)
Jiao M, He W, Ouyang Z, Qin Q, Guo Y, Zhang J, Bai Y, Guo X, Yu Q, She J, Hwang PM, Zheng F, Wen Y
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRN9_fit1_model1.png)
|
| Sample: |
Polysorbate 20 (PS20) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 0, 45 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 2
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.4 |
nm |
| Dmax |
8.6 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRP9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of Polysorbate 20 (PS20) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 0, 45 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 2
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
8.8 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRQ9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan monolaurate fraction (F2) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 20-mer, 24 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.0 |
nm |
| Dmax |
8.0 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRR9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan monolaurate fraction (F2) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 22-mer, 26 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Mar 6
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.1 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRS9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan higher order esters fraction (F4) with no or low amount of the fatty acid myristic acid (MA) (< 100 µg/ml) 0, 42 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 29
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.3 |
nm |
|
|
|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDRT9_fit1_model1.png)
|
| Sample: |
Polydisperse core-shell ellipsoidal micelles of POE sorbitan higher order esters fraction (F4) with high amount of the fatty acid myristic acid (MA) (> 500 µg/ml) 0, 42 kDa
|
| Buffer: |
aqueous solution of 4% methanol, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 29
|
Small-angle x-ray scattering investigation of the integration of free fatty acids in polysorbate 20 micelles
Biophysical Journal (2023)
Ehrit J, Gräwert T, Göddeke H, Konarev P, Svergun D, Nagel N
|
| RgGuinier |
3.5 |
nm |
| Dmax |
8.7 |
nm |
|
|
|
|
|
|
|
| Sample: |
Recombination protein 107 tetramer, 139 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
8.3 |
nm |
| Dmax |
32.2 |
nm |
|
|
|
|
|
|

|
| Sample: |
WGS project CABT00000000 data, contig 2.12 tetramer, 155 kDa Sordaria macrospora (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
9.6 |
nm |
| Dmax |
38.0 |
nm |
|
|
|
|
|
|

|
| Sample: |
Protein PAIR1 tetramer, 132 kDa Zea mays protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
8.0 |
nm |
| Dmax |
31.7 |
nm |
|
|