|
|
|
|
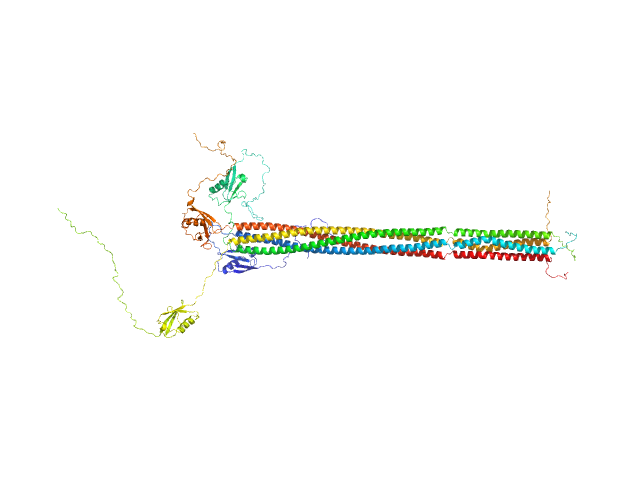
|
| Sample: |
Putative recombination initiation defects 3 tetramer, 129 kDa Arabidopsis thaliana protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
7.8 |
nm |
| Dmax |
27.5 |
nm |
|
|
|
|
|
|

|
| Sample: |
Meiotic recombination protein rec15 tetramer, 128 kDa Schizosaccharomyces pombe (strain … protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
6.9 |
nm |
| Dmax |
26.5 |
nm |
|
|
|
|
|
|
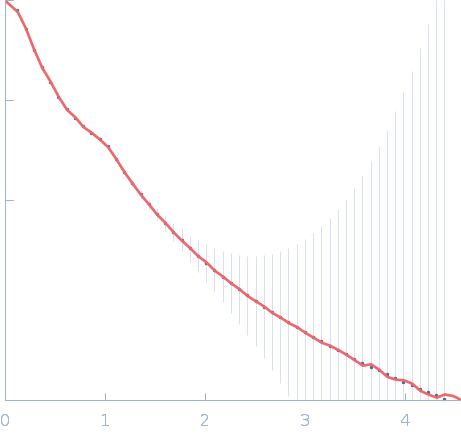
|
| Sample: |
Interactor of HORMAD1 protein 1 (M198V, V230I) tetramer, 127 kDa Mus musculus protein
|
| Buffer: |
25 mM HEPES-NaOH, 500 mM NaCl, 5 mM EDTA, 5% glycerol, 1 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 14
|
Evolutionary conservation of the structure and function of meiotic Rec114-Mei4 and Mer2 complexes.
Genes Dev 37(11-12):535-553 (2023)
Daccache D, De Jonge E, Liloku P, Mechleb K, Haddad M, Corthaut S, Sterckx YG, Volkov AN, Claeys Bouuaert C
|
| RgGuinier |
7.2 |
nm |
| Dmax |
25.5 |
nm |
|
|
|
|
|
|
|
| Sample: |
DUF2285 domain-containing protein monomer, 11 kDa Mesorhizobium loti protein
|
| Buffer: |
100 mM NaCl, 50 mM Tris-HCl, 2% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 1
|
DUF2285 is a novel helix-turn-helix domain variant that orchestrates both activation and antiactivation of conjugative element transfer in proteobacteria.
Nucleic Acids Res (2023)
Jowsey WJ, Morris CRP, Hall DA, Sullivan JT, Fagerlund RD, Eto KY, Solomon PD, Mackay JP, Bond CS, Ramsay JP, Ronson CW
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.7 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
At3g55760 monomer, 60 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 8
|
LIKE EARLY STARVATION 1 and EARLY STARVATION 1 promote and stabilize amylopectin phase transition in starch biosynthesis.
Sci Adv 9(21):eadg7448 (2023)
Liu C, Pfister B, Osman R, Ritter M, Heutinck A, Sharma M, Eicke S, Fischer-Stettler M, Seung D, Bompard C, Abt MR, Zeeman SC
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
121 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Inactive purple acid phosphatase-like protein monomer, 40 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 150 mM NaCl, 10% glycerol, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2016 Dec 8
|
LIKE EARLY STARVATION 1 and EARLY STARVATION 1 promote and stabilize amylopectin phase transition in starch biosynthesis.
Sci Adv 9(21):eadg7448 (2023)
Liu C, Pfister B, Osman R, Ritter M, Heutinck A, Sharma M, Eicke S, Fischer-Stettler M, Seung D, Bompard C, Abt MR, Zeeman SC
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
83 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CP12-2 (D77G) monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
25 mM potassium phosphate, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Conformational Disorder Analysis of the Conditionally Disordered Protein CP12 from Arabidopsis thaliana in Its Different Redox States
International Journal of Molecular Sciences 24(11):9308 (2023)
Del Giudice A, Gurrieri L, Galantini L, Fanti S, Trost P, Sparla F, Fermani S
|
| RgGuinier |
2.0 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
10 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
CP12-2 (D77G) monomer, 8 kDa Arabidopsis thaliana protein
|
| Buffer: |
25 mM potassium phosphate, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Feb 15
|
Conformational Disorder Analysis of the Conditionally Disordered Protein CP12 from Arabidopsis thaliana in Its Different Redox States
International Journal of Molecular Sciences 24(11):9308 (2023)
Del Giudice A, Gurrieri L, Galantini L, Fanti S, Trost P, Sparla F, Fermani S
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.6 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
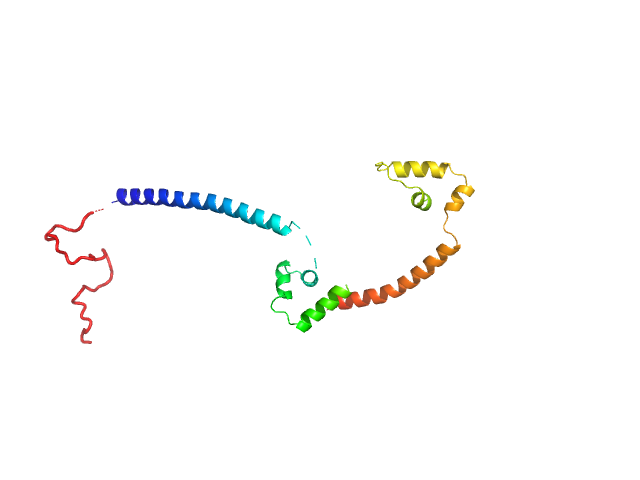
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
|
| Buffer: |
50mM Tris, 750mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2017 Oct 10
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.7 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Gcf1p monomer, 26 kDa Candida albicans (strain … protein
Af2_20 DNA monomer, 12 kDa DNA
|
| Buffer: |
25 mM Tris, 20 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Aug 30
|
Structural analysis of the Candida albicans mitochondrial DNA maintenance factor Gcf1p reveals a dynamic DNA-bridging mechanism.
Nucleic Acids Res (2023)
Tarrés-Solé A, Battistini F, Gerhold JM, Piétrement O, Martínez-García B, Ruiz-López E, Lyonnais S, Bernadó P, Roca J, Orozco M, Le Cam E, Sedman J, Solà M
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|