|
|
|
|
|
| Sample: |
Non-structural protein V monomer, 9 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
2.8 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V (ΔC-terminal and Y111A, Y112A, Y113A mutant) monomer, 9 kDa Hendra virus (isolate … protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
2.7 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
21 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Non-structural protein V monomer, 15 kDa Nipah henipavirus protein
|
| Buffer: |
50 mM sodium phosphate, pH: 7.2
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2022 Jul 14
|
Molecular Determinants of Fibrillation in a Viral Amyloidogenic Domain from Combined Biochemical and Biophysical Studies
International Journal of Molecular Sciences 24(1):399 (2022)
Nilsson J, Baroudi H, Gondelaud F, Pesce G, Bignon C, Ptchelkine D, Chamieh J, Cottet H, Kajava A, Longhi S
|
| RgGuinier |
3.7 |
nm |
| Dmax |
16.5 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Fusion protein of LSm and MyoX-coil , 1066 kDa Artificial protein protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 5% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 May 23
|
Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J Am Chem Soc (2022)
Ohara N, Kawakami N, Arai R, Adachi N, Moriya T, Kawasaki M, Miyamoto K
|
| RgGuinier |
9.1 |
nm |
| Dmax |
22.7 |
nm |
|
|
|
|
|
|

|
| Sample: |
TIP60 (K67E) mutant (metal-ion induced 60-mer complex with barium ions) , 1066 kDa Artificial protein protein
Barium ion 0, 8 kDa
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 5% glycerol, 5 mM BaCl2, pH: 8
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 May 23
|
Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J Am Chem Soc (2022)
Ohara N, Kawakami N, Arai R, Adachi N, Moriya T, Kawasaki M, Miyamoto K
|
| RgGuinier |
9.6 |
nm |
| Dmax |
21.8 |
nm |
|
|
|
|
|
|

|
| Sample: |
TIP60 (K67E) mutant with EDTA dimer, 36 kDa Artificial protein protein
|
| Buffer: |
25 mM HEPES, 100 mM NaCl, 1 mM EDTA, 5% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2021 May 23
|
Reversible Assembly of an Artificial Protein Nanocage Using Alkaline Earth Metal Ions.
J Am Chem Soc (2022)
Ohara N, Kawakami N, Arai R, Adachi N, Moriya T, Kawasaki M, Miyamoto K
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
|
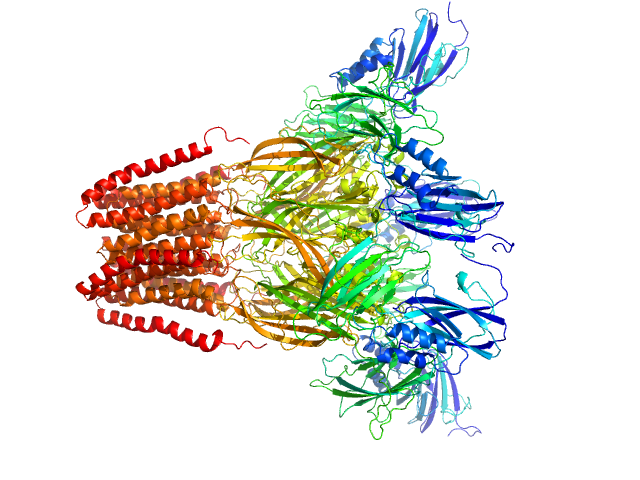
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 10 mM CaCl2, 0.5 mM deuterated n-Dodecyl-B-D-Maltoside, in D2O, pH: 7.5
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 23
|
Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc Natl Acad Sci U S A 119(50):e2210669119 (2022)
Lycksell M, Rovšnik U, Hanke A, Martel A, Howard RJ, Lindahl E
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
530 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Neurotransmitter-gated ion-channel ligand-binding domain-containing protein pentamer, 342 kDa Desulfofustis sp. PB-SRB1 protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 10 mM EDTA, 0.5 mM deuterated n-Dodecyl-B-D-Maltoside, in D2O, pH: 7.5
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2020 Aug 23
|
Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc Natl Acad Sci U S A 119(50):e2210669119 (2022)
Lycksell M, Rovšnik U, Hanke A, Martel A, Howard RJ, Lindahl E
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
540 |
nm3 |
|
|
|
|
|
|
|
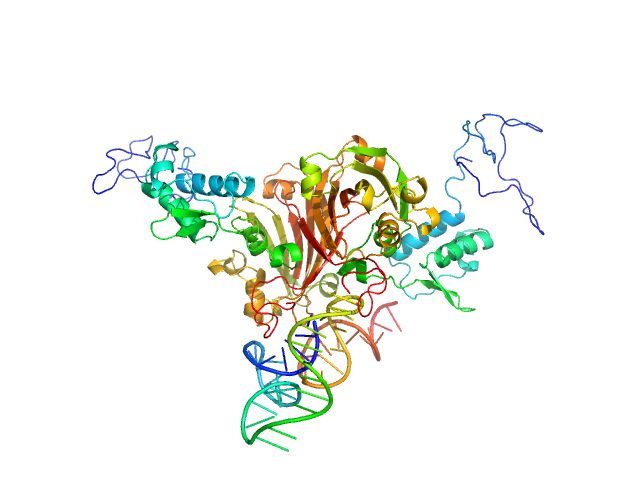
| Sample: |
Complex of Rv0792c and Rv0792c_1 monomer, 74 kDa
|
| Buffer: |
25 mM HEPES Buffer; 400 mM NaCl,, pH: 7.2
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 31
|
Structural and Functional Characterization of Rv0792c from Mycobacterium tuberculosis: Identifying Small Molecule Inhibitor against HutC Protein.
Microbiol Spectr :e0197322 (2022)
Chauhan NK, Anand A, Sharma A, Dhiman K, Gosain TP, Singh P, Singh P, Khan E, Chattopadhyay G, Kumar A, Sharma D, Ashish, Sharma TK, Singh R
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
|
|
|
|
|
|
|
| Sample: |
Complex of Rv0792c and Rv0792c_2 monomer, 46 kDa
|
| Buffer: |
25 mM HEPES Buffer; 400 mM NaCl,, pH: 7.2
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 31
|
Structural and Functional Characterization of Rv0792c from Mycobacterium tuberculosis: Identifying Small Molecule Inhibitor against HutC Protein.
Microbiol Spectr :e0197322 (2022)
Chauhan NK, Anand A, Sharma A, Dhiman K, Gosain TP, Singh P, Singh P, Khan E, Chattopadhyay G, Kumar A, Sharma D, Ashish, Sharma TK, Singh R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.8 |
nm |
|
|