|
|
|
|
|
| Sample: |
Replicase polyprotein 1ab dimer, 63 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 500 mM NaCl, 5% glycerol, and 1 mM TCEP, pH: 8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2020 Oct 15
|
Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro.
Sci Adv 8(49):eadd2191 (2022)
Yadav R, Courouble VV, Dey SK, Harrison JJEK, Timm J, Hopkins JB, Slack RL, Sarafianos SG, Ruiz FX, Griffin PR, Arnold E
|
| RgGuinier |
3.3 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
110 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Replicase polyprotein 1a monomer, 60 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES, 10% glycerol, 500 mM NaCl, 5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro.
Sci Adv 8(49):eadd2191 (2022)
Yadav R, Courouble VV, Dey SK, Harrison JJEK, Timm J, Hopkins JB, Slack RL, Sarafianos SG, Ruiz FX, Griffin PR, Arnold E
|
| RgGuinier |
3.5 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1a dimer, 119 kDa Severe acute respiratory … protein
|
| Buffer: |
20 mM HEPES, 10% glycerol, 500 mM NaCl, 5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2021 Nov 14
|
Biochemical and structural insights into SARS-CoV-2 polyprotein processing by Mpro.
Sci Adv 8(49):eadd2191 (2022)
Yadav R, Courouble VV, Dey SK, Harrison JJEK, Timm J, Hopkins JB, Slack RL, Sarafianos SG, Ruiz FX, Griffin PR, Arnold E
|
| RgGuinier |
4.6 |
nm |
| Dmax |
19.1 |
nm |
| VolumePorod |
203 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
von Willebrand factor C6 domain wild type monomer, 9 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 11
|
Structure and dynamics of the von Willebrand Factor C6 domain
Journal of Structural Biology 214(4):107923 (2022)
Chen P, Kutzki F, Mojzisch A, Simon B, Xu E, Aponte-Santamaría C, Horny K, Jeffries C, Schneppenheim R, Wilmanns M, Brehm M, Gräter F, Hennig J
|
| RgGuinier |
1.7 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspB monomer, 48 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 21
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
5.0 |
nm |
| Dmax |
19.2 |
nm |
| VolumePorod |
119 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspB monomer, 37 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 21
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
75 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspB monomer, 30 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 21
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
3.3 |
nm |
| Dmax |
10.8 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
|
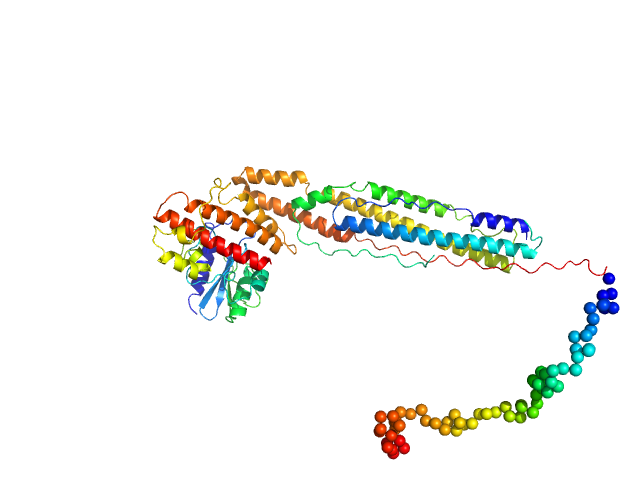
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
ESX-1 secretion-associated protein EspB monomer, 48 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 12
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
5.0 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
128 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
ESX-1 secretion-associated protein EspB monomer, 37 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Apr 12
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.7 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspK monomer, 27 kDa Mycobacterium tuberculosis (strain … protein
ESX-1 secretion-associated protein EspB monomer, 30 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 12
|
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
4.1 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
113 |
nm3 |
|
|