|
|
|
|

|
| Sample: |
Multidrug resistance operon repressor dimer, 32 kDa Pseudomonas aeruginosa protein
|
| Buffer: |
20mM HEPES, 150mM NaCl, 10mM DTT, 1% v/v glycerol, pH: 7.1
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Nov 23
|
Small-angle X-ray and neutron scattering of MexR and its complex with DNA supports a conformational selection binding model
Biophysical Journal (2022)
Caporaletti F, Pietras Z, Morad V, Mårtensson L, Gabel F, Wallner B, Martel A, Sunnerhagen M
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.7 |
nm |
| VolumePorod |
56 |
nm3 |
|
|
|
|
|
|
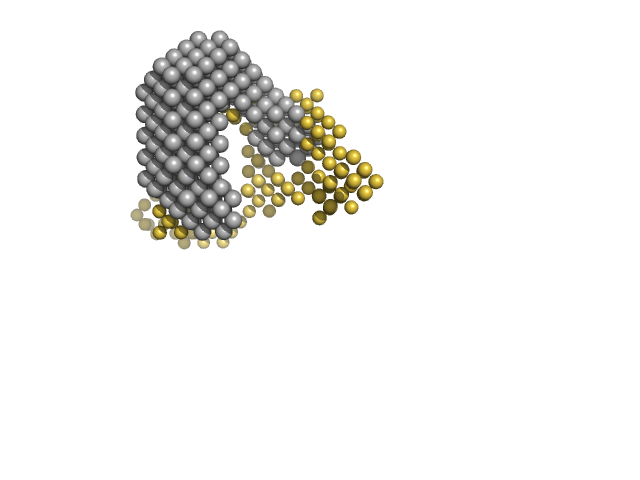
|
| Sample: |
Multidrug resistance operon repressor dimer, 32 kDa Pseudomonas aeruginosa protein
34 base pair double-stranded DNA monomer, 21 kDa synthetic construct DNA
|
| Buffer: |
20mM NaPO4, 150 mM NaCl, 10 mM DTT, pH: 7.1
|
| Experiment: |
SANS
data collected at D22, Institut Laue-Langevin (ILL) on 2018 May 30
|
Small-angle X-ray and neutron scattering of MexR and its complex with DNA supports a conformational selection binding model
Biophysical Journal (2022)
Caporaletti F, Pietras Z, Morad V, Mårtensson L, Gabel F, Wallner B, Martel A, Sunnerhagen M
|
| RgGuinier |
2.9 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
ESX-1 secretion-associated protein EspB heptamer, 261 kDa Mycobacterium tuberculosis (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 300 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Sep 12
|
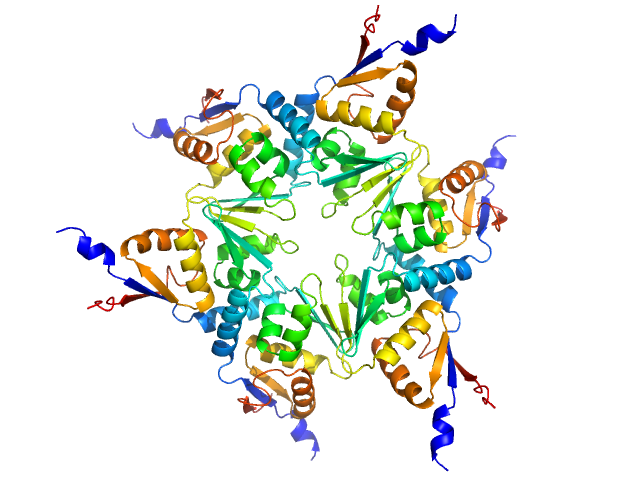
The crystal structure of the EspB-EspK virulence factor-chaperone complex suggests an additional type VII secretion mechanism in M. tuberculosis.
J Biol Chem :102761 (2022)
Gijsbers A, Eymery M, Gao Y, Menart I, Vinciauskaite V, Siliqi D, Peters PJ, McCarthy A, Ravelli RBG
|
| RgGuinier |
5.9 |
nm |
| Dmax |
18.8 |
nm |
| VolumePorod |
911 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Longitudinals lacking protein, isoform G hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
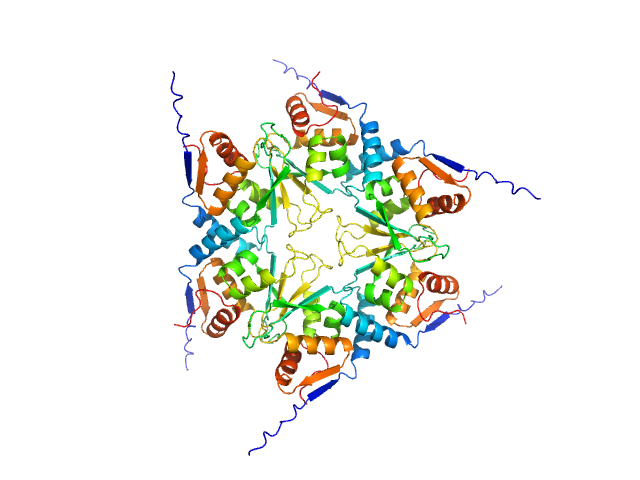
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Longitudinals lacking protein, isoform G hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.8 |
nm |
| Dmax |
24.0 |
nm |
| VolumePorod |
263 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
167 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Jul 8
|
BTB domains: A structural view of evolution, multimerization, and protein-protein interactions.
Bioessays :e2200179 (2022)
Bonchuk A, Balagurov K, Georgiev P
|
| RgGuinier |
4.1 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
192 |
nm3 |
|
|
|
|
|
|
|
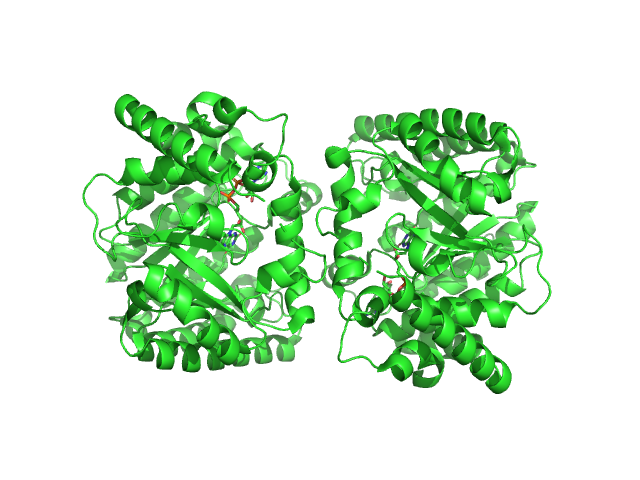
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
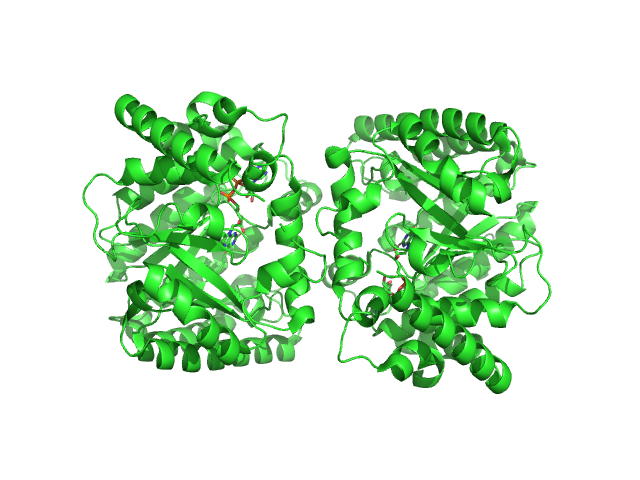
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
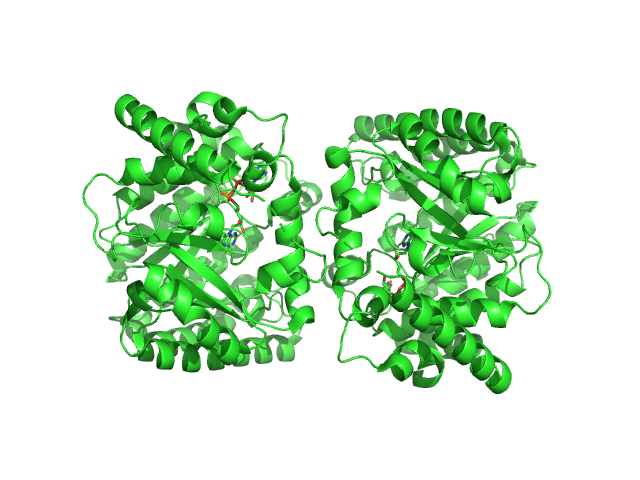
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.7 |
nm |
| VolumePorod |
102 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Minimal proline dehydrogenase domain of proline utilization A (design #2) dimer, 87 kDa Sinorhizobium meliloti protein
|
| Buffer: |
25 mM HEPES pH 7.6, 150 mM NaCl, and 1mM TCEP, pH: 7.6
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2022 Apr 12
|
Structure-based engineering of minimal Proline dehydrogenase domains for inhibitor discovery.
Protein Eng Des Sel (2022)
Bogner AN, Ji J, Tanner JJ
|
| RgGuinier |
3.0 |
nm |
| Dmax |
9.8 |
nm |
| VolumePorod |
108 |
nm3 |
|
|