|
|
|
|
|
| Sample: |
Lipid None, lipid
|
| Buffer: |
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jul 25
|
Lipid_unpublished_73
Siawosch Schewa
|
|
|
|
|
|
|
|
| Sample: |
Lipid None, lipid
|
| Buffer: |
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 18
|
Lipid_unpublished_75
Siawosch Schewa
|
|
|
|
|
|
|
|
| Sample: |
Lipid None, lipid
|
| Buffer: |
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Jul 25
|
Lipid_unpublished_74
Siawosch Schewa
|
|
|
|
|
|
|
|
| Sample: |
Lipid None, lipid
|
| Buffer: |
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 19
|
Lipid_unpublished_76
Siawosch Schewa
|
|
|
|
|
|
|
|
| Sample: |
Lipid None, lipid
|
| Buffer: |
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Dec 19
|
Lipid_unpublished_77
Siawosch Schewa
|
|
|
|
|
|
|
|
| Sample: |
Toll-like receptor dimer, 175 kDa Aedes aegypti protein
|
| Buffer: |
50 mM NaCl, 50 mM Tris-HCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Aug 2
|
Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun 13(1):5110 (2022)
Saucereau Y, Wilson TH, Tang MCK, Moncrieffe MC, Hardwick SW, Chirgadze DY, Soares SG, Marcaida MJ, Gay NJ, Gangloff M
|
| RgGuinier |
5.4 |
nm |
| Dmax |
20.5 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Toll-like receptor dimer, 175 kDa Aedes aegypti protein
AAEL013433-PA (Cystine-knot_cytokine) dimer, 26 kDa Aedes aegypti protein
|
| Buffer: |
50 mM NaCl, 50 mM Tris-HCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Aug 2
|
Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aegypti.
Nat Commun 13(1):5110 (2022)
Saucereau Y, Wilson TH, Tang MCK, Moncrieffe MC, Hardwick SW, Chirgadze DY, Soares SG, Marcaida MJ, Gay NJ, Gangloff M
|
| RgGuinier |
5.5 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
567 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nominal 100 nm diameter polystyrene spheres monomer, 315312 kDa
|
| Buffer: |
Water, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 16
|
Standard polystyrene nanospheres (concentration series data)
Dima Molodenskiy
|
| RgGuinier |
33.0 |
nm |
| Dmax |
100.0 |
nm |
| VolumePorod |
520 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nominal 20 nm diameter polystyrene spheres 0, 8 kDa
|
| Buffer: |
Water, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 May 16
|
Standard polystyrene nanospheres (concentration series data)
Dima Molodenskiy
|
| RgGuinier |
8.5 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
3 |
nm3 |
|
|
|
|
|
|
|
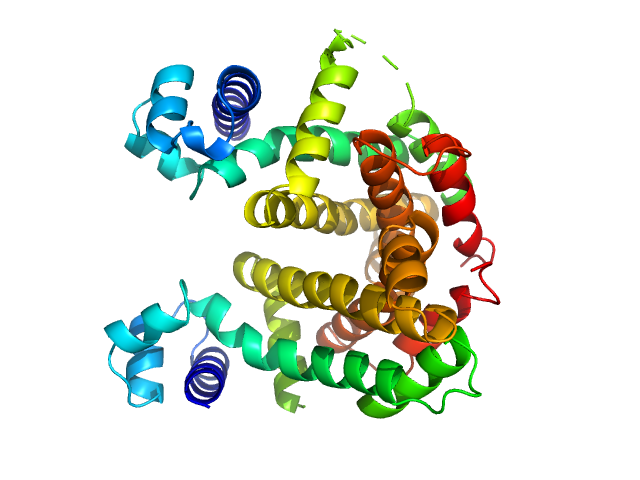
| Sample: |
Putative tetR-family transcriptional regulator dimer, 47 kDa Streptomyces coelicolor (strain … protein
|
| Buffer: |
20 mM Tris/HCl, 400 mM NaCl, 50 mM imidazole, 1 mM DTT, pH: 8.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2014 May 2
|
Crystal Structures of Free and Ligand-Bound Forms of the TetR/AcrR-Like Regulator SCO3201 from Streptomyces coelicolor Suggest a Novel Allosteric Mechanism.
FEBS J (2022)
Werten S, Waack P, Palm GJ, Virolle MJ, Hinrichs W
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
77 |
nm3 |
|
|