|
|
|
|
|
| Sample: |
β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5
|
| Experiment: |
SANS
data collected at D11, ILL on 2019 Sep 21
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
|
|
|
|
|
| Sample: |
β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5
|
| Experiment: |
SANS
data collected at D11, ILL on 2019 Sep 21
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
|
|
|
|
|
| Sample: |
β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM Acetate, 72% v/v D₂O, pH: 5
|
| Experiment: |
SANS
data collected at D11, ILL on 2019 Sep 21
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
|
|
|
|
|
| Sample: |
β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5
|
| Experiment: |
SANS
data collected at D11, ILL on 2019 Sep 21
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
|
|
|
|
|
| Sample: |
β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5
|
| Experiment: |
SANS
data collected at D11, ILL on 2020 Aug 18
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
|
|
|
|
|
| Sample: |
β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
GlcNAc-binding protein A monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5
|
| Experiment: |
SANS
data collected at D11, ILL on 2020 Aug 18
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
|
|
|
|
|
| Sample: |
β-chitin nanofibers from squid pens None, Squid
GlcNAc-binding protein A (perdeuterated) monomer, 51 kDa Vibrio cholerae serotype … protein
GlcNAc-binding protein A monomer, 51 kDa Vibrio cholerae serotype … protein
|
| Buffer: |
20 mM acetate, 47% v/v D₂O, pH: 5
|
| Experiment: |
SANS
data collected at D11, ILL on 2020 Aug 18
|
Tangled up in fibers: How a lytic polysaccharide monooxygenase binds its chitin substrate
Henrik Vinther Sørensen
|
|
|
|
|
|
|
|
| Sample: |
NAC domain-containing protein 13 dimer, 63 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM NaNO3, 5 mM TCEP, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 3
|
Allovalent scavenging of activation domains in the transcription factor ANAC013 gears transcriptional regulation.
Nucleic Acids Res 53(4) (2025)
Delaforge E, Due AD, Theisen FF, Morffy N, O'Shea C, Blackledge M, Strader LC, Skriver K, Kragelund BB
|
| RgGuinier |
3.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
118 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
NAC domain-containing protein 13 monomer, 13 kDa Arabidopsis thaliana protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 5 mM NaNO3, 5 mM TCEP, pH: 7.4
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2022 Oct 3
|
Allovalent scavenging of activation domains in the transcription factor ANAC013 gears transcriptional regulation.
Nucleic Acids Res 53(4) (2025)
Delaforge E, Due AD, Theisen FF, Morffy N, O'Shea C, Blackledge M, Strader LC, Skriver K, Kragelund BB
|
| RgGuinier |
3.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
36 |
nm3 |
|
|
|
|
|
|
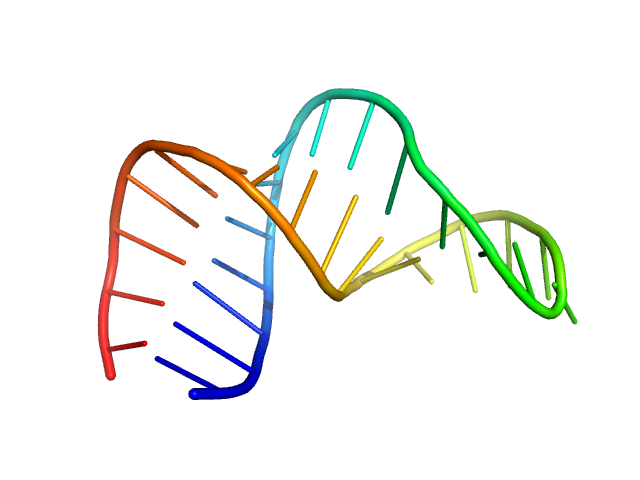
|
| Sample: |
HIV-1 dimerization initiation site with a CCCCCC apical loop monomer, 9 kDa RNA
|
| Buffer: |
50 mM potassium phosphate buffer, 1 mM MgCl2, 50 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Feb 20
|
Selective deuteration of an RNA:RNA complex for structural analysis using small-angle scattering.
Structure (2025)
Munsayac A, Leite WC, Hopkins JB, Hall I, O'Neill HM, Keane SC
|
| RgGuinier |
1.5 |
nm |
| Dmax |
4.6 |
nm |
| VolumePorod |
11 |
nm3 |
|
|