|
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDML5_electron_density.png)
|
| Sample: |
Proteoliposomes composed of lipids from A/Puerto Rico/8/34 (H1N1) virus envelope together with the HA LI45 peptides loaded with M1 protein (lipid:M1 molar ration 4:1) monomer, 2000 kDa
|
| Buffer: |
100 mM NaCl, 50 mM MES buffer, pH: 6.8
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 26
|
The Cytoplasmic Tail of Influenza A Virus Hemagglutinin and Membrane Lipid Composition Change the Mode of M1 Protein Association with the Lipid Bilayer
Membranes 11(10):772 (2021)
Kordyukova L, Konarev P, Fedorova N, Shtykova E, Ksenofontov A, Loshkarev N, Dadinova L, Timofeeva T, Abramchuk S, Moiseenko A, Baratova L, Svergun D, Batishchev O
|
|
|
|
|
|
|
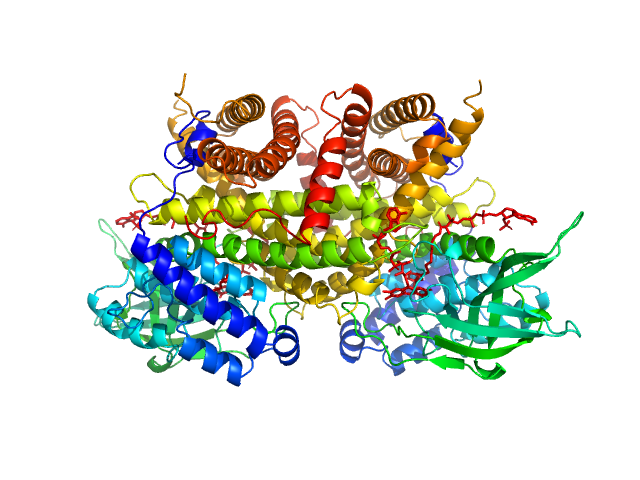
|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 130 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
213 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial dimer, 130 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial dimer, 93 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.8 |
nm |
| VolumePorod |
514 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Complex I assembly factor ACAD9, mitochondrial tetramer, 259 kDa Homo sapiens protein
Evolutionarily conserved signaling intermediate in Toll pathway, mitochondrial tetramer, 184 kDa Homo sapiens protein
Complex I intermediate-associated protein 30, mitochondrial tetramer, 190 kDa Homo sapiens protein
|
| Buffer: |
25mM Tris-HCl, 150mM NaCl, 0.1mM DTT, and 5% glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2017 Nov 2
|
Molecular mechanism of interactions between ACAD9 and binding partners in mitochondrial respiratory complex I assembly
iScience 24(10):103153 (2021)
Xia C, Lou B, Fu Z, Mohsen A, Shen A, Vockley J, Kim J
|
| RgGuinier |
6.6 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
1340 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Coronin tetramer, 21 kDa Trypanosoma brucei brucei … protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR - Institute of Microbial Technology (IMTech) on 2019 Oct 17
|
Structural insights into kinetoplastid coronin oligomerization domain and F-actin interaction
Current Research in Structural Biology (2021)
Parihar P, Singh A, Karade S, Sahasrabuddhe A, Pratap J
|
| RgGuinier |
3.3 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cell cycle associated protein MOB1, putative monomer, 27 kDa Leishmania donovani (strain … protein
|
| Buffer: |
100 mM Tris-HCl, 100 mM NaCl, pH: 7.4
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2017 Jun 22
|
Structural insights into kinetoplastid coronin oligomerization domain and F-actin interaction
Current Research in Structural Biology (2021)
Parihar P, Singh A, Karade S, Sahasrabuddhe A, Pratap J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
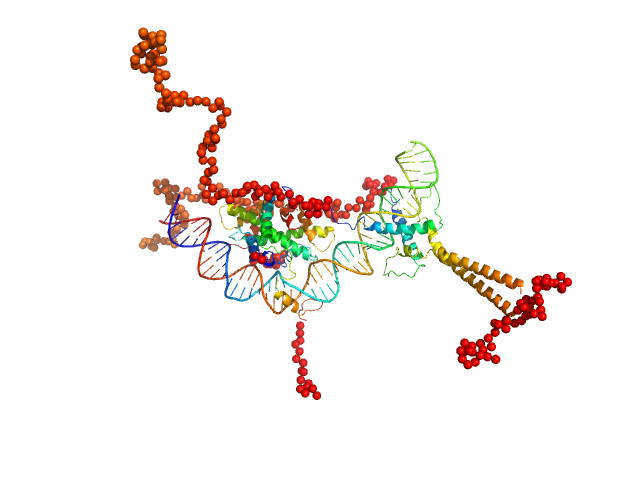
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
155 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Upstream stimulatory factor 1 dimer, 50 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 50bp monomer, 31 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2015 Jun 14
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
4.8 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Protein max (Isoform 2, short, 13-21: missing) dimer, 39 kDa Homo sapiens protein
Nuclear transcription factor Y subunit alpha monomer, 10 kDa Homo sapiens protein
Nuclear transcription factor Y subunit beta monomer, 11 kDa Homo sapiens protein
Nuclear transcription factor Y subunit gamma monomer, 11 kDa Homo sapiens protein
DNA 48bp monomer, 30 kDa DNA
|
| Buffer: |
100 mM cacodylate buffer, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2012 Dec 10
|
The USR domain of USF1 mediates NF-Y interactions and cooperative DNA binding
International Journal of Biological Macromolecules (2021)
Bernardini A, Lorenzo M, Chaves-Sanjuan A, Swuec P, Pigni M, Saad D, Konarev P, Graewert M, Valentini E, Svergun D, Nardini M, Mantovani R, Gnesutta N
|
| RgGuinier |
5.2 |
nm |
| Dmax |
17.0 |
nm |
| VolumePorod |
151 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclic di-AMP synthase CdaA dimer, 44 kDa Bacillus subtilis (strain … protein
|
| Buffer: |
30 mM Tris, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Jul 21
|
Structural basis for the inhibition of the Bacillus subtilis c-di-AMP cyclase CdaA by the phosphoglucomutase GlmM
Journal of Biological Chemistry :101317 (2021)
Pathania M, Tosi T, Millership C, Hoshiga F, Morgan R, Freemont P, Gründling A
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
60 |
nm3 |
|
|