|
|
|
|
|
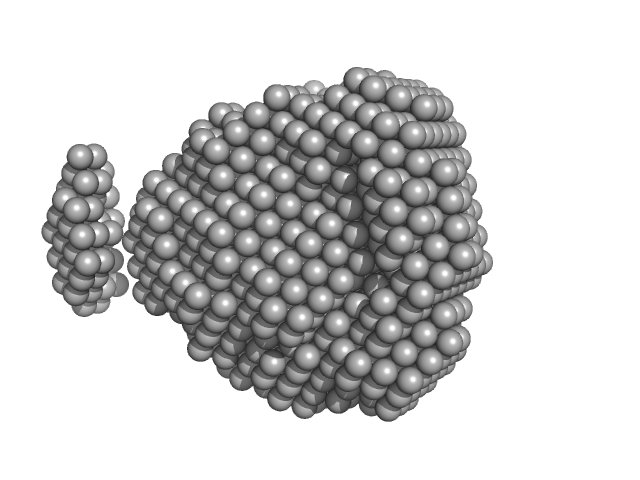
| Sample: |
Fructokinase, PfkB monomer, 32 kDa Mycobacterium marinum (strain … protein
|
| Buffer: |
20 mM Tris-HCl, 100 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2019 Dec 17
|
Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum
Biochemical and Biophysical Research Communications (2021)
Gao B, Ji R, Li Z, Su X, Li H, Sun Y, Ji C, Gan J, Li J
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.6 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein tetramer, 79 kDa Menangle virus protein
|
| Buffer: |
12.5 mM MOPS/KOH pH 7.0, 250 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Nov 16
|
Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions
Viruses 13(9):1737 (2021)
Webby M, Herr N, Bulloch E, Schmitz M, Keown J, Goldstone D, Kingston R
|
| RgGuinier |
6.3 |
nm |
| Dmax |
23.4 |
nm |
| VolumePorod |
524 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein monomer, 13 kDa Menangle virus protein
|
| Buffer: |
12.5 mM MOPS/KOH pH 7.0, 150 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2016 Aug 17
|
Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions
Viruses 13(9):1737 (2021)
Webby M, Herr N, Bulloch E, Schmitz M, Keown J, Goldstone D, Kingston R
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Phosphoprotein monomer, 6 kDa Menangle virus protein
|
| Buffer: |
12.5 mM Tris/HCl pH 8.5, 150 mM NaCl, pH: 8.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Aug 15
|
Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions
Viruses 13(9):1737 (2021)
Webby M, Herr N, Bulloch E, Schmitz M, Keown J, Goldstone D, Kingston R
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.3 |
nm |
| VolumePorod |
12 |
nm3 |
|
|
|
|
|
|
|
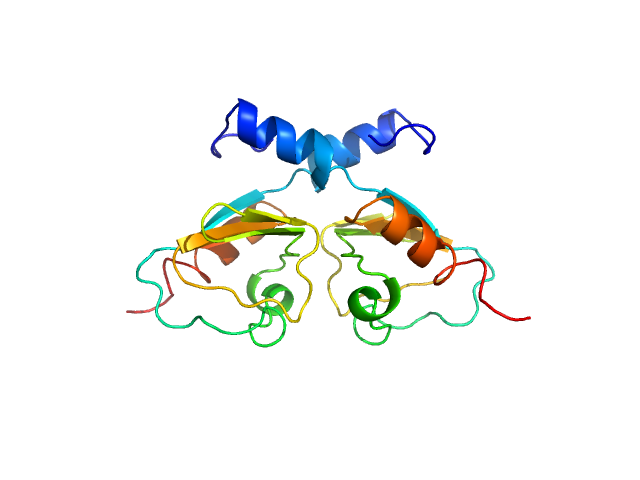
| Sample: |
Transcriptional repressor BusR RCK_C domain dimer, 22 kDa Streptococcus agalactiae serotype … protein
|
| Buffer: |
100 mM NaCl, 30mM Hepes, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
1.9 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
44 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Transcriptional repressor BusR tetramer, 95 kDa Streptococcus agalactiae protein
|
| Buffer: |
20mM HEPES, pH6.5, 100mM NaCl, 3% glycerol (v/v), pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
4.4 |
nm |
| Dmax |
13.9 |
nm |
| VolumePorod |
168 |
nm3 |
|
|
|
|
|
|
|
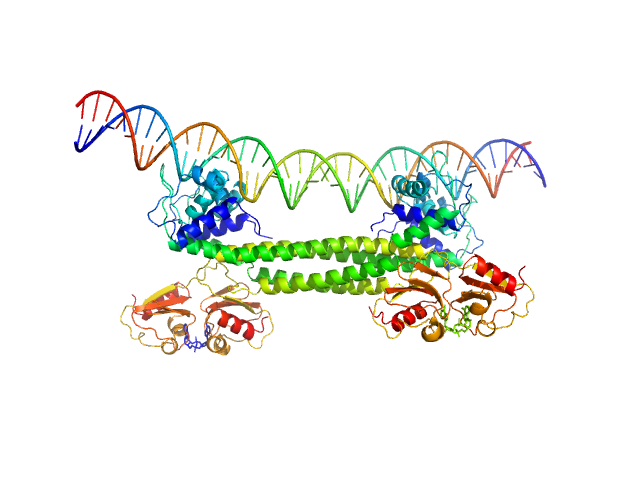
| Sample: |
Transcriptional repressor BusR tetramer, 95 kDa Streptococcus agalactiae protein
BusR Recognition sequence monomer, 28 kDa synthetic construct DNA
|
| Buffer: |
20mM HEPES, pH6.5, 100mM NaCl, 3% glycerol (v/v), pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Jul 2
|
BusR senses bipartite DNA binding motifs by a unique molecular ruler architecture.
Nucleic Acids Res (2021)
Bandera AM, Bartho J, Lammens K, Drexler DJ, Kleinschwärzer J, Hopfner KP, Witte G
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
210 |
nm3 |
|
|
|
|
|
|
|
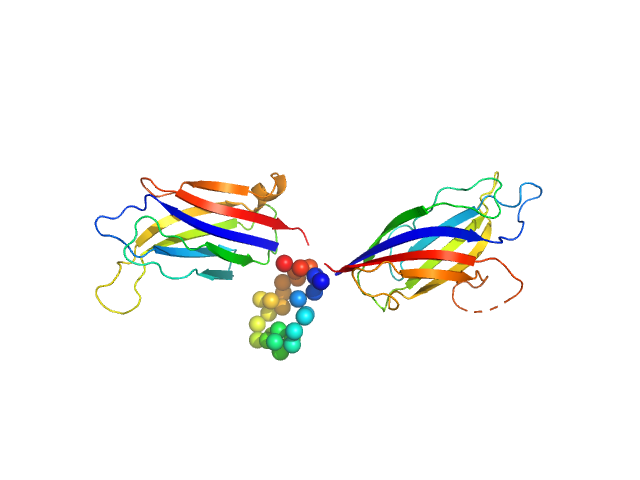
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jun 11
|
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.1 |
nm |
| VolumePorod |
59 |
nm3 |
|
|
|
|
|
|
|
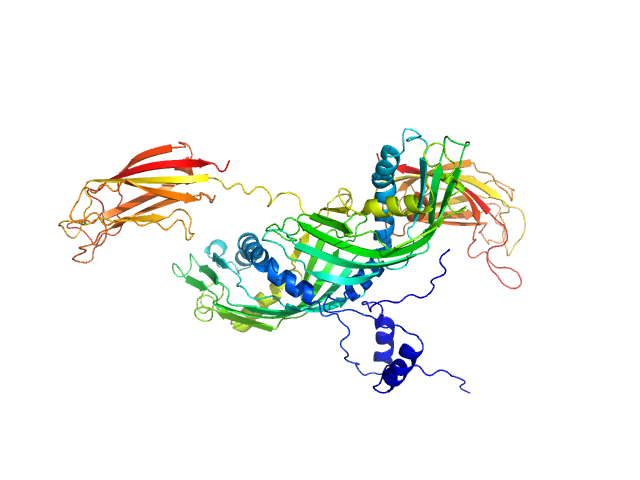
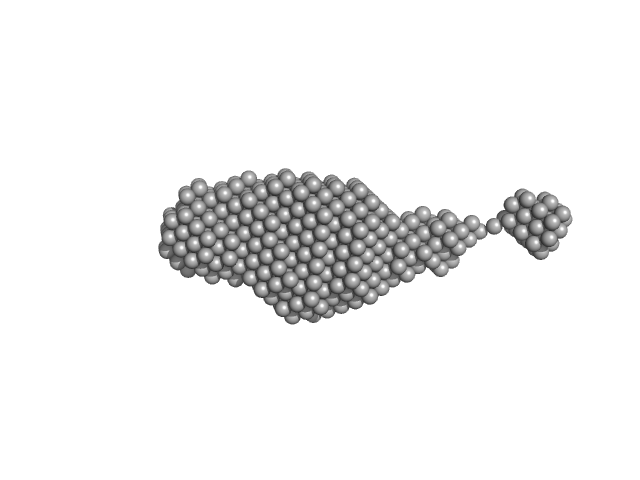
| Sample: |
Synaptotagmin-1 (SYT1-SMP2C2A) monomer, 72 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, 5% glycerol, 2 mM DTT, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2020 Jun 11
|
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
4.2 |
nm |
| Dmax |
17.8 |
nm |
| VolumePorod |
138 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Synaptotagmin-1 monomer, 33 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM Tris, 50 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2021 Feb 16
|
The structure and flexibility analysis of the Arabidopsis
synaptotagmin 1 reveal the basis of its regulation at membrane contact sites
Life Science Alliance 4(10):e202101152 (2021)
Benavente J, Siliqi D, Infantes L, Lagartera L, Mills A, Gago F, Ruiz-López N, Botella M, Sánchez-Barrena M, Albert A
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
50 |
nm3 |
|
|