|
|
|
|
|
| Sample: |
Plasmodium falciparum Heat shock protein 90 N-terminal and Middle domains monomer, 68 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Plasmodium falciparum Heat shock protein 90 middle domain monomer, 33 kDa Plasmodium falciparum protein
|
| Buffer: |
25 mM Tris-HCl, 100 mM KCl, 1 mM β-mercaptoethanol, 1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS1 Beamline, Brazilian Synchrotron Light Laboratory on 2018 May 11
|
Solution structure of Plasmodium falciparum Hsp90 indicates a high flexible dimer
Archives of Biochemistry and Biophysics :108468 (2020)
Silva N, Torricillas M, Minari K, Barbosa L, Seraphim T, Borges J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
47 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD)) monomer, 23 kDa Hathewaya histolytica protein
|
| Buffer: |
10 mM HEPES, 100 mM NaCl, 0.4 mM EGTA, 2.4 mM CaCl2, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2015 Sep 25
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in...
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.7 |
nm |
| VolumePorod |
29 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cell wall synthesis protein Wag31 tetramer, 34 kDa Mycobacterium tuberculosis protein
|
| Buffer: |
20 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2018 Sep 3
|
Structural basis of self-assembly in the lipid-binding domain of mycobacterial polar growth factor Wag31
IUCrJ 7(4):767-776 (2020)
Choukate K, Chaudhuri B
|
| RgGuinier |
2.7 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
52 |
nm3 |
|
|
|
|
|
|

|
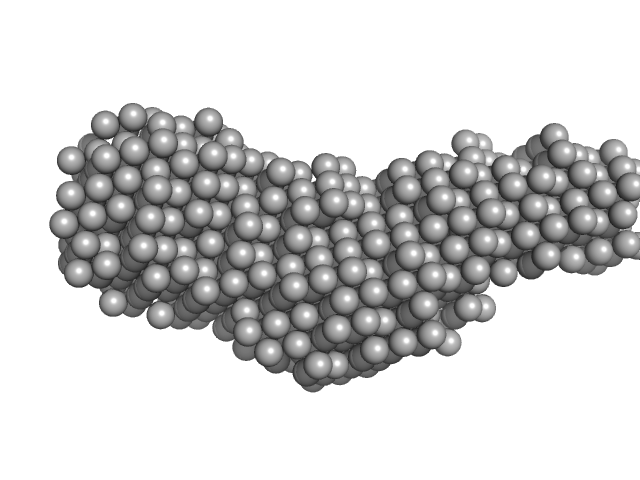
| Sample: |
Pomacea diffusa perivitellin-1 , 23 kDa Pomacea diffusa protein
|
| Buffer: |
50 mM Phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at SAXS2 Beamline, Brazilian Synchrotron Light Laboratory on 2015 Mar 27
|
A highly stable, non-digestible lectin from Pomacea diffusa unveils clade-related protection systems in apple snail eggs.
J Exp Biol 223(Pt 19) (2020)
Brola TR, Dreon MS, Qiu JW, Heras H
|
| RgGuinier |
4.6 |
nm |
| Dmax |
15.5 |
nm |
| VolumePorod |
529 |
nm3 |
|
|
|
|
|
|
|
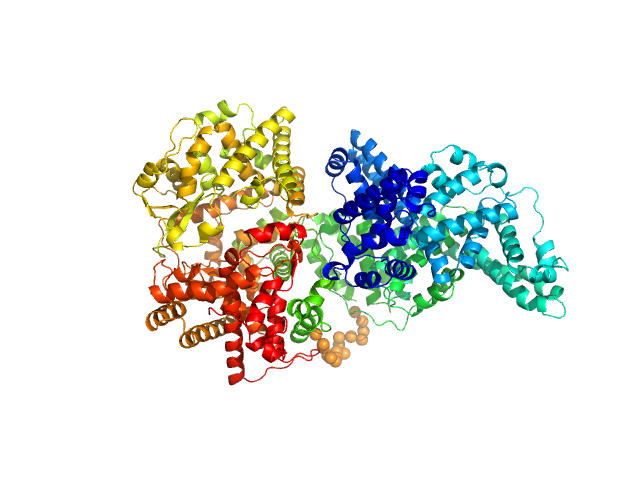
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM histidine, pH: 5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 21
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
267 |
nm3 |
|
|
|
|
|
|
|
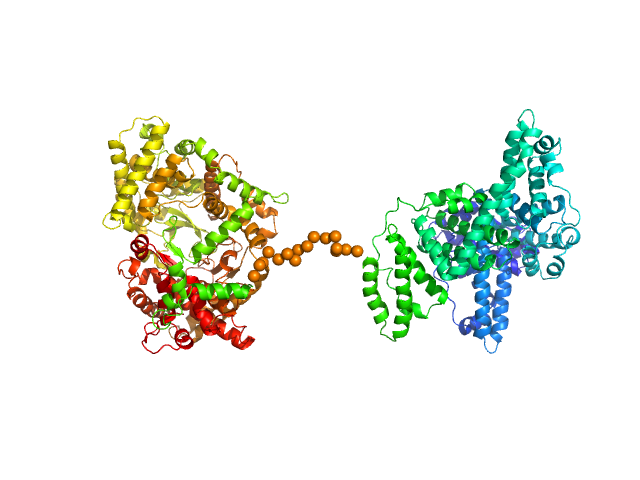
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM histidine, pH: 5.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 Dec 21
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
274 |
nm3 |
|
|
|
|
|
|
|
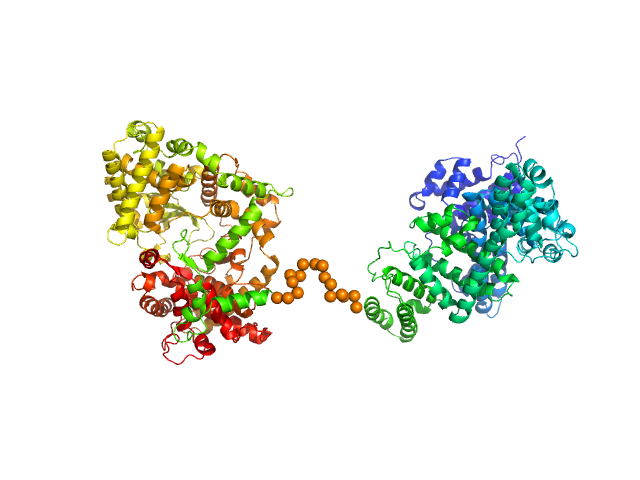
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM histidine, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 7
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.6 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
258 |
nm3 |
|
|
|
|
|
|
|
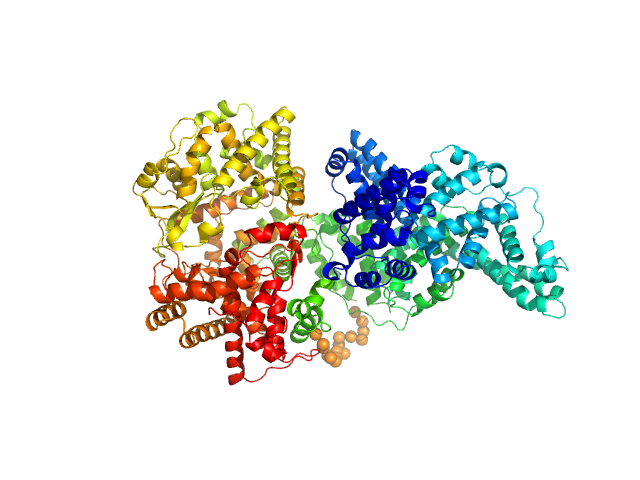
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM histidine, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2017 Jun 26
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
5.0 |
nm |
| Dmax |
17.4 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
|
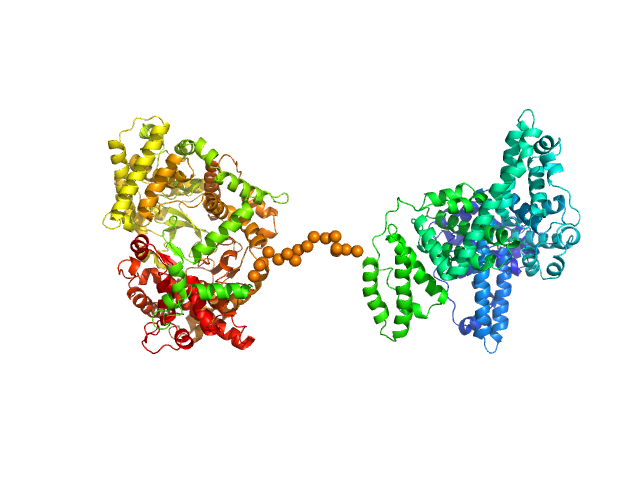
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM TRIS, pH: 8.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Dec 15
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
239 |
nm3 |
|
|