|
|
|
|
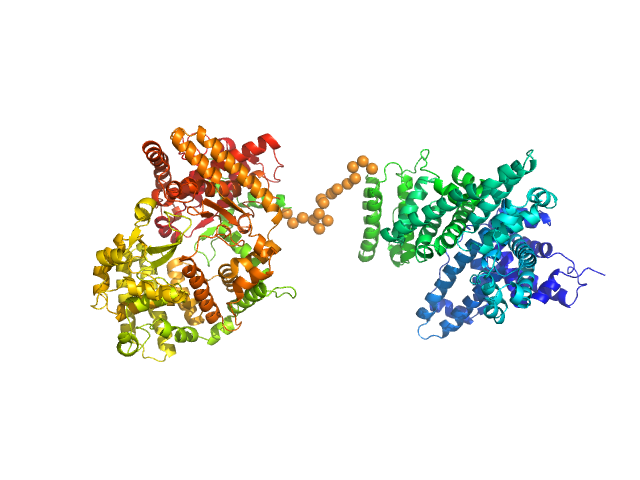
|
| Sample: |
Neprilysin - G400V mutant monomer, 80 kDa Homo sapiens protein
Human serum albumin - C58S mutant monomer, 66 kDa Homo sapiens protein
|
| Buffer: |
10 mM phosphate, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Jul 7
|
Albumin-neprilysin fusion protein: understanding stability using small angle X-ray scattering and molecular dynamic simulations.
Sci Rep 10(1):10089 (2020)
Kulakova A, Indrakumar S, Sønderby Tuelung P, Mahapatra S, Streicher WW, Peters GHJ, Harris P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
16.7 |
nm |
| VolumePorod |
240 |
nm3 |
|
|
|
|
|
|
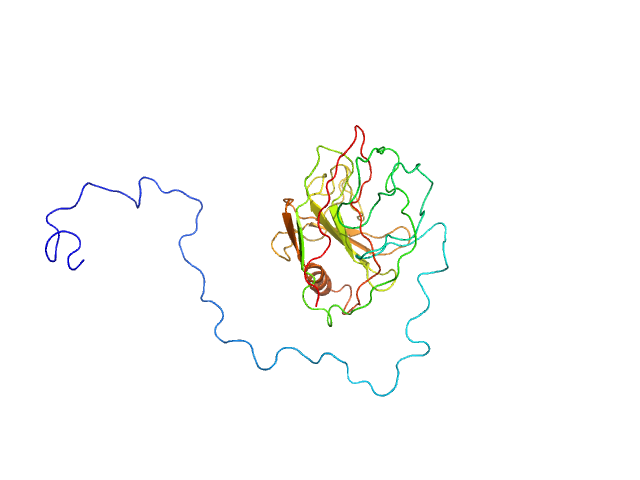
|
| Sample: |
Type III effector NopAA monomer, 31 kDa Sinorhizobium fredii USDA257 protein
|
| Buffer: |
PBS, 150 mM NaCl, 10% glycerol, pH: 7.4
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2014 Dec 17
|
Structural and enzymatic characterisation of the Type III effector NopAA (=GunA) from Sinorhizobium fredii USDA257 reveals a Xyloglucan hydrolase activity.
Sci Rep 10(1):9932 (2020)
Dorival J, Philys S, Giuntini E, Brailly R, de Ruyck J, Czjzek M, Biondi E, Bompard C
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 2
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
94 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly(rC)-binding protein 2 monomer, 40 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 16
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.3 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly(rC)-binding protein 2 monomer, 40 kDa Homo sapiens protein
modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Dec 16
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
162 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.8 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
165 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Truncated poly(rC)-binding protein 2 (ΔKH3) monomer, 28 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jan 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
66 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
modified stem loop IV poliovirus IRES, nucleotides 278-398 monomer, 41 kDa Human poliovirus 1 … RNA
Truncated poly(rC)-binding protein 2 (ΔKH1-KH2) monomer, 18 kDa Homo sapiens protein
|
| Buffer: |
5 mM HEPES-KOH, 25 mM KCl, 2 mM MgCl2, 2 mM DTT, 4 % glycerol, 0.1 mM EDTA, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jul 1
|
Structure of the PCBP2/stem-loop IV complex underlying translation initiation mediated by the poliovirus type I IRES.
Nucleic Acids Res (2020)
Beckham SA, Matak MY, Belousoff MJ, Venugopal H, Shah N, Vankadari N, Elmlund H, Nguyen JHC, Semler BL, Wilce MCJ, Wilce JA
|
| RgGuinier |
3.5 |
nm |
| Dmax |
12.2 |
nm |
| VolumePorod |
126 |
nm3 |
|
|
|
|
|
|
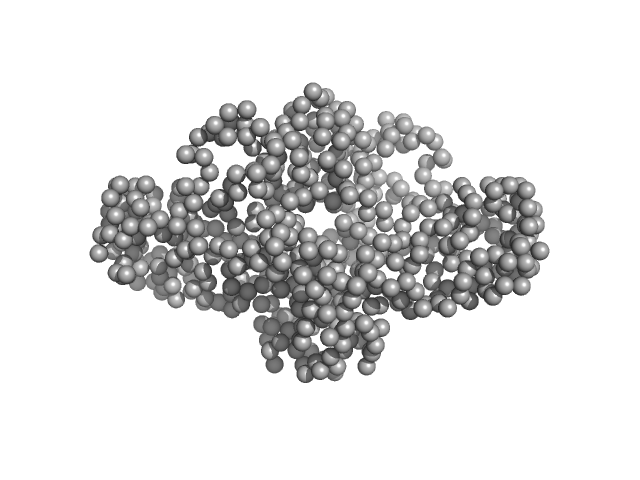
|
| Sample: |
Uncharacterized protein dimer, 66 kDa Legionella pneumophila protein
|
| Buffer: |
20 mM Tris, 200 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2019 Sep 18
|
Structure, Dynamics and Cellular Insight Into Novel Substrates of the Legionella pneumophila Type II Secretion System
Frontiers in Molecular Biosciences 7 (2020)
Portlock T, Tyson J, Dantu S, Rehman S, White R, McIntire I, Sewell L, Richardson K, Shaw R, Pandini A, Cianciotto N, Garnett J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
|
|
|
|
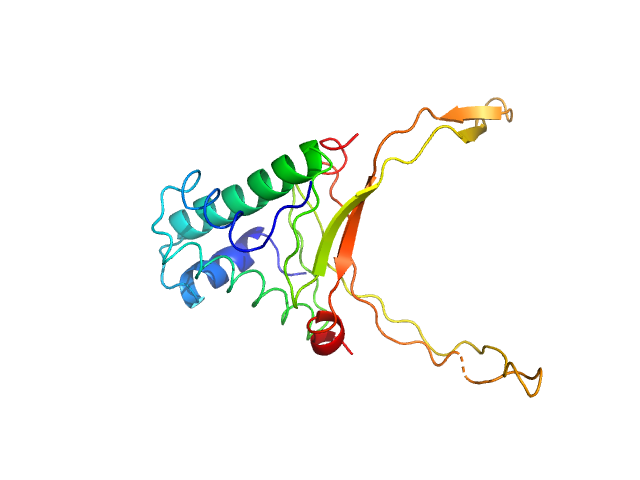
|
| Sample: |
DNA-binding protein HU-alpha octamer, 77 kDa Escherichia coli protein
|
| Buffer: |
10 mM Bis-Tris, 100 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 May 27
|
Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun 11(1):2905 (2020)
Remesh SG, Verma SC, Chen JH, Ekman AA, Larabell CA, Adhya S, Hammel M
|
| RgGuinier |
3.2 |
nm |
| Dmax |
10.7 |
nm |
|
|