|
|
|
|
|
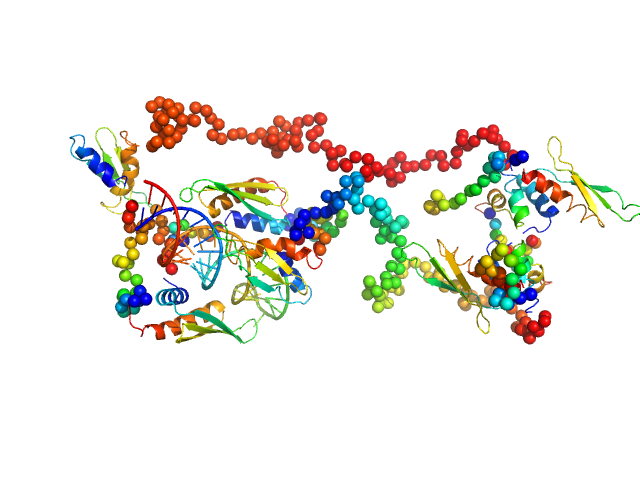
| Sample: |
Group 1 truncated hemoglobin (C51S, C71S, Y108A) , 13 kDa Shewanella benthica KT99 protein
|
| Buffer: |
14 mM Tris, 6 mM potassium phosphate, pH: 7
|
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Feb 18
|
Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N
Journal of Biological Chemistry :108223 (2025)
Martinez Grundman J, Schultz T, Schlessman J, Johnson E, Gillilan R, Lecomte J
|
| RgGuinier |
2.3 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
31 |
nm3 |
|
|
|
|
|
|
|
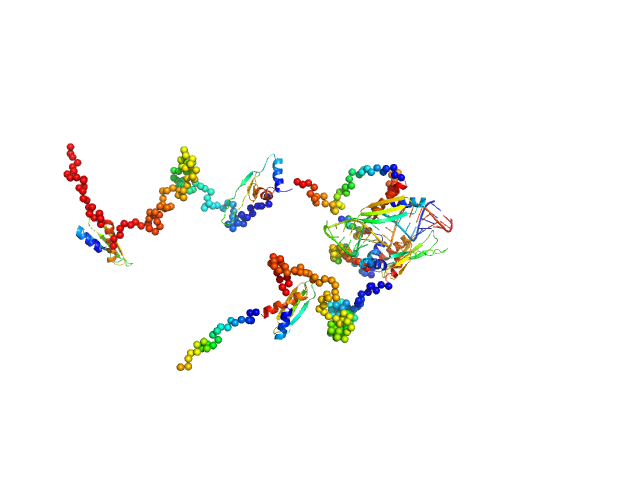
| Sample: |
Double-stranded RNA-binding protein Staufen homolog 1 (Δ1-177) dimer, 89 kDa Homo sapiens protein
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-1000, CEITEC on 2020 May 4
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
4.9 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
129 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
3'UTR fragment of ADP-ribosylation factor 1 monomer, 14 kDa Homo sapiens RNA
Double-stranded RNA-binding protein Staufen homolog 1 with truncated RNA-binding domain 2 and truncated Staufen-swapping (ΔSSM) dimer, 81 kDa Homo sapiens protein
|
| Buffer: |
50 mM TRIS, 300 mM NaCl, 3.8 mM β-mercaptoethanol, pH: 7
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, CEITEC on 2024 Jan 12
|
A Simple Protocol for Visualization of RNA-Protein Complexes by Atomic Force Microscopy.
Curr Protoc 5(1):e70084 (2025)
Tripepi A, Shakoor H, Klapetek P
|
| RgGuinier |
5.5 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
139 |
nm3 |
|
|
|
|
|
|
|
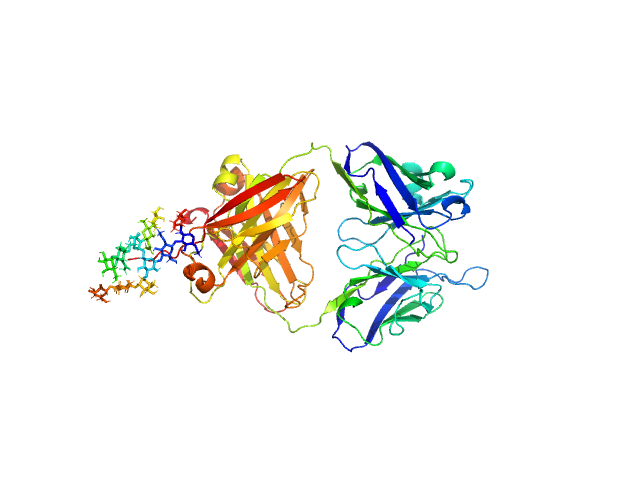
| Sample: |
IgM Mannitou Fab Heavy Chain monomer, 26 kDa Mus musculus protein
IgM Mannitou Fab Light Chain monomer, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 300 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2024 Jun 5
|
Small-angle X-ray scattering of engineered antigen-binding fragments: the case of glycosylated Fab from the Mannitou IgM antibody.
Acta Crystallogr F Struct Biol Commun (2025)
Semwal S, Karamolegkou M, Flament S, Raouraoua N, Verstraete K, Thureau A, Wien F, Bray F, Savvides SN, Bouckaert J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Alpha/beta fold hydrolase monomer, 28 kDa Staphylococcus aureus protein
|
| Buffer: |
100 mM NaCl, 10 mM HEPES, pH: 7.6
|
| Experiment: |
SAXS
data collected at BioSAXS, Australian Synchrotron on 2024 Apr 11
|
Similar but Distinct—Biochemical Characterization of the
Staphylococcus aureus
Serine Hydrolases FphH
and FphI
Proteins: Structure, Function, and Bioinformatics (2024)
Fellner M, Randall G, Bitac I, Warrender A, Sethi A, Jelinek R, Kass I
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 (L24A) monomer, 23 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 160 mM NaCl, 0.5 mM TCEP pH 7.5, pH: 7.5
|
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Dec 9
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
21.6 |
nm |
| VolumePorod |
127 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 160 mM NaCl, 0.5 mM TCEP pH 7.5, pH: 7.5
|
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2023 Dec 9
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
4.6 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
236 |
nm3 |
|
|
|
|
|
|
|
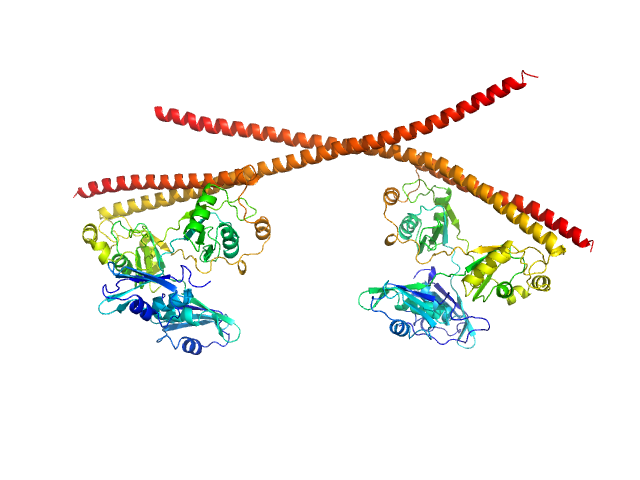
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 500 mM NaCl, 0.5 mM TCEP, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 11
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
5.5 |
nm |
| Dmax |
25.4 |
nm |
| VolumePorod |
171 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Partner and localizer of BRCA2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 160 mM NaCl, 0.5 mM TCEP, pH: 7.4
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 11
|
The strand exchange domain of tumor suppressor PALB2 is intrinsically disordered and promotes oligomerization-dependent DNA compaction.
iScience 27(12):111259 (2024)
Kyriukha Y, Watkins MB, Redington JM, Chintalapati N, Ganti A, Dastvan R, Uversky VN, Hopkins JB, Pozzi N, Korolev S
|
| RgGuinier |
4.7 |
nm |
| Dmax |
21.3 |
nm |
| VolumePorod |
197 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Splicing factor, proline- and glutamine-rich dimer, 153 kDa Homo sapiens protein
Non-POU domain-containing octamer-binding protein dimer, 60 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris pH 7.5, 250 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Jun 18
|
Structural plasticity of the coiled-coil interactions in human SFPQ.
Nucleic Acids Res (2024)
Koning HJ, Lai JY, Marshall AC, Stroeher E, Monahan G, Pullakhandam A, Knott GJ, Ryan TM, Fox AH, Whitten A, Lee M, Bond CS
|
| RgGuinier |
5.5 |
nm |
| Dmax |
20.4 |
nm |
| VolumePorod |
304 |
nm3 |
|
|