|
|
|
|
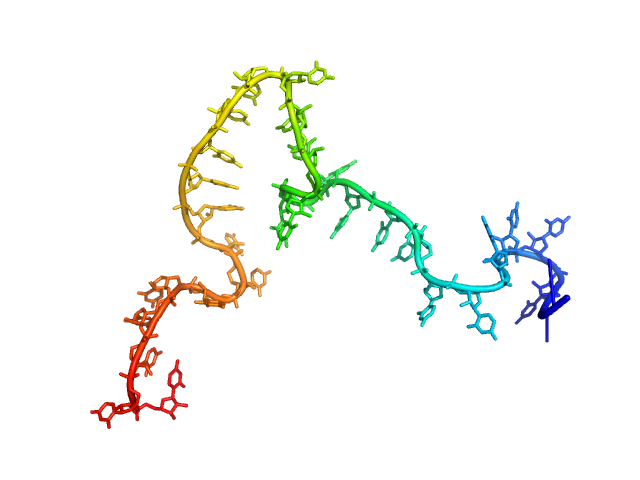
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 1 mM MgCl2, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.7 |
nm |
| Dmax |
11.0 |
nm |
| VolumePorod |
17 |
nm3 |
|
|
|
|
|
|
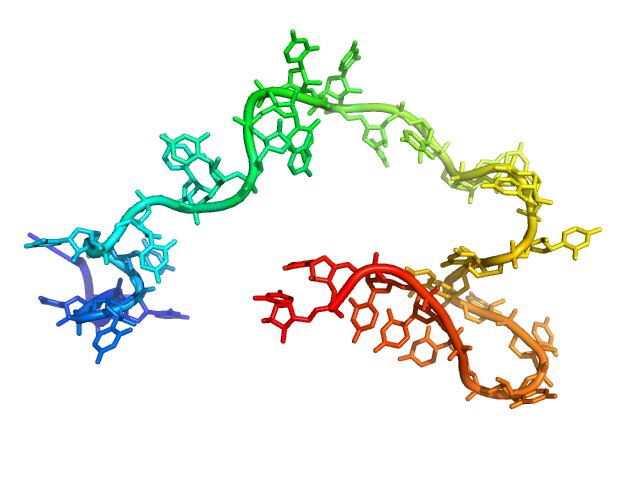
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 2 mM MgCl2, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.6 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 5 mM MgCl2, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.5 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
15 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Poly-uridine monomer, 9 kDa RNA
|
| Buffer: |
1 mM Na-MOPS, 20 mM NaCl, 10 mM MgCl2, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2015 Oct 24
|
Visualizing disordered single-stranded RNA: connecting sequence, structure and electrostatics.
J Am Chem Soc (2019)
Plumridge A, Andresen K, Pollack L
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
14 |
nm3 |
|
|
|
|
|
|
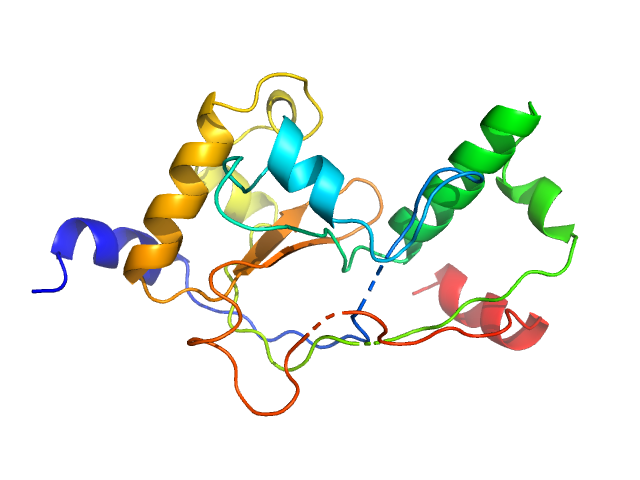
|
| Sample: |
Mycobacterial cidal toxin monomer, 20 kDa Mycobacterium tuberculosis H37Rv protein
|
| Buffer: |
30 mM Tris-HCl, 200 mM NaCl, 10% glycerol, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2017 May 2
|
MbsTA
Diana Freire
|
| RgGuinier |
1.8 |
nm |
| Dmax |
5.6 |
nm |
| VolumePorod |
30 |
nm3 |
|
|
|
|
|
|
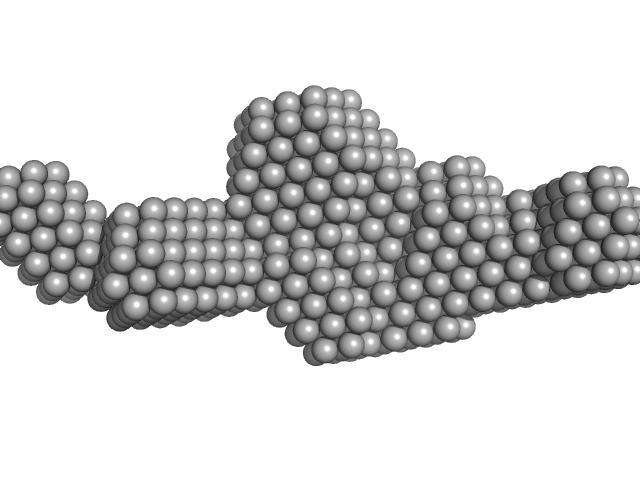
|
| Sample: |
Sa0446 binding sequence 40bp monomer, 25 kDa DNA
Transcriptional regulator Lrs14-like protein dimer, 33 kDa Sulfolobus acidocaldarius protein
|
| Buffer: |
300 mM NaCl, 20 mM HEPES, pH 7.5, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Nov 5
|
Solution Structure of Archaeal Biofilm Regulator 2 (AbfR2) in Complex with 40 bp DNA
Marian Vogt
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Endoribonuclease E tetramer, 247 kDa Escherichia coli protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.0 |
nm |
| Dmax |
16.1 |
nm |
| VolumePorod |
468 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 248 kDa Yersinia pestis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.1 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
470 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 256 kDa Francisella tularensis protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
5.1 |
nm |
| Dmax |
17.2 |
nm |
| VolumePorod |
491 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Endoribonuclease E tetramer, 250 kDa Burkholderia pseudomallei protein
|
| Buffer: |
10 mM DTT, 10 mM MgCl2, 0.5 M NaCl, 20 mM Tris, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Feb 11
|
A structural and biochemical comparison of Ribonuclease E homologues from pathogenic bacteria highlights species-specific properties.
Sci Rep 9(1):7952 (2019)
Mardle CE, Shakespeare TJ, Butt LE, Goddard LR, Gowers DM, Atkins HS, Vincent HA, Callaghan AJ
|
| RgGuinier |
4.8 |
nm |
| Dmax |
14.9 |
nm |
| VolumePorod |
437 |
nm3 |
|
|