|
|
|
|
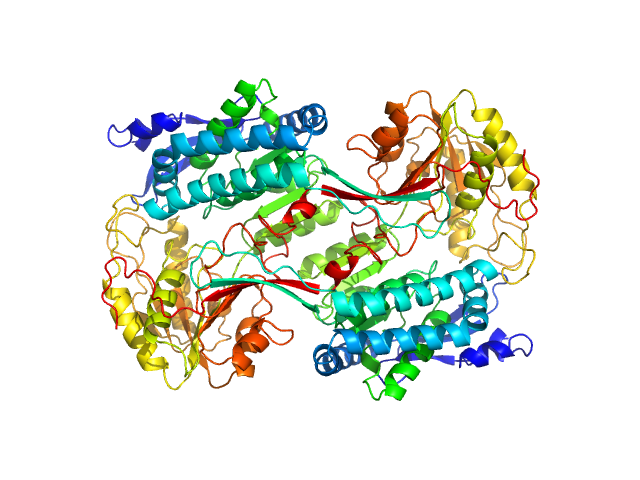
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399Q , 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
255 |
nm3 |
|
|
|
|
|
|
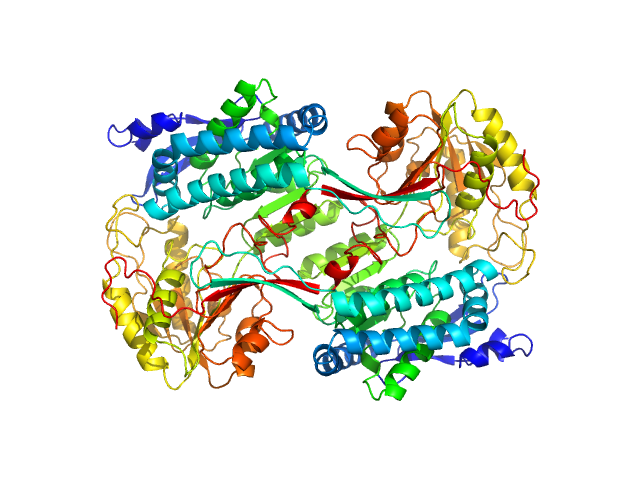
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase , 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
256 |
nm3 |
|
|
|
|
|
|
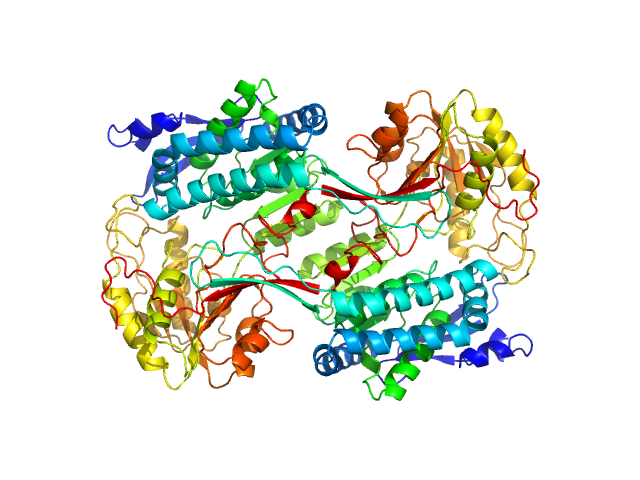
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase , 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.4 |
nm |
| VolumePorod |
260 |
nm3 |
|
|
|
|
|
|
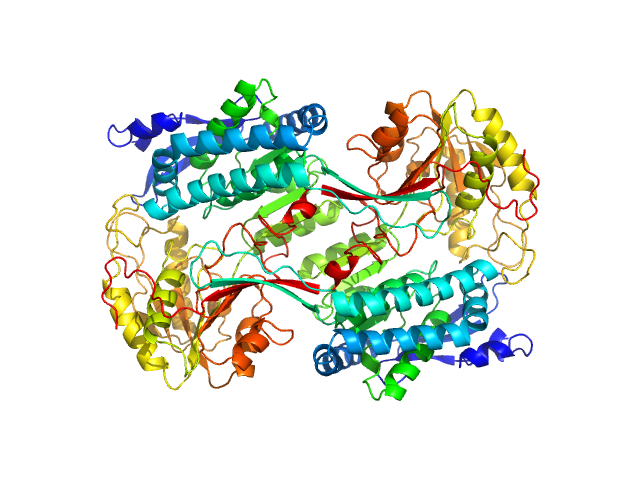
|
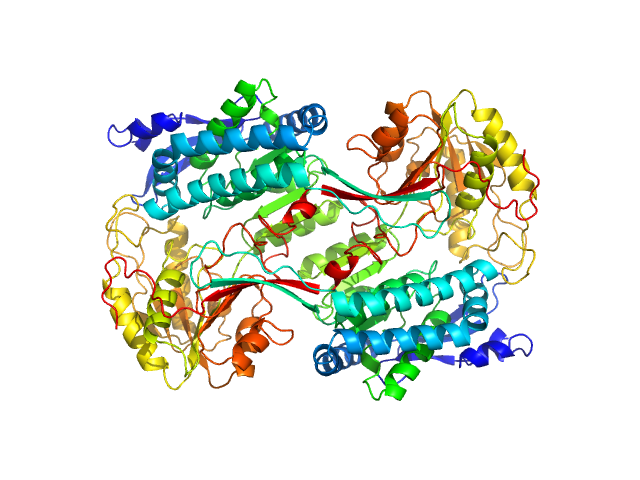
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399D , 56 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.9 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
272 |
nm3 |
|
|
|
|
|
|
|
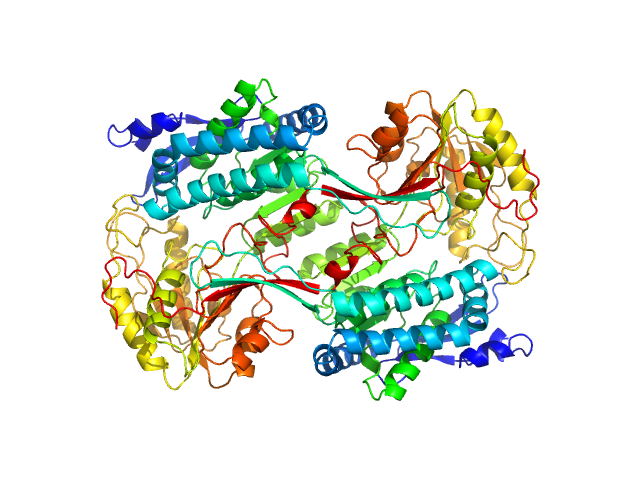
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399G , 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
11.3 |
nm |
| VolumePorod |
290 |
nm3 |
|
|
|
|
|
|
|
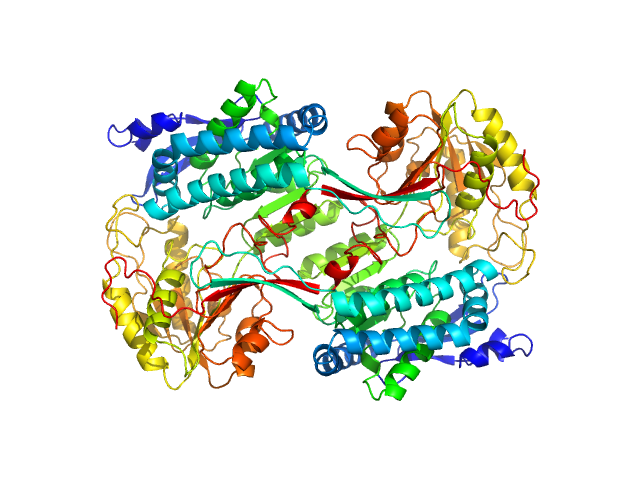
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399G , 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.7 |
nm |
| Dmax |
10.6 |
nm |
| VolumePorod |
250 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Alpha-aminoadipic semialdehyde dehydrogenase E399G , 55 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 1 mM DTT, 10 mM NAD, 2% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 May 1
|
Structural Analysis of Pathogenic Mutations Targeting Glu427 of ALDH7A1, the Hot Spot Residue of Pyridoxine-Dependent Epilepsy.
J Inherit Metab Dis (2019)
Laciak AR, Korasick DA, Gates KS, Tanner JJ
|
| RgGuinier |
3.8 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
270 |
nm3 |
|
|
|
|
|
|
|
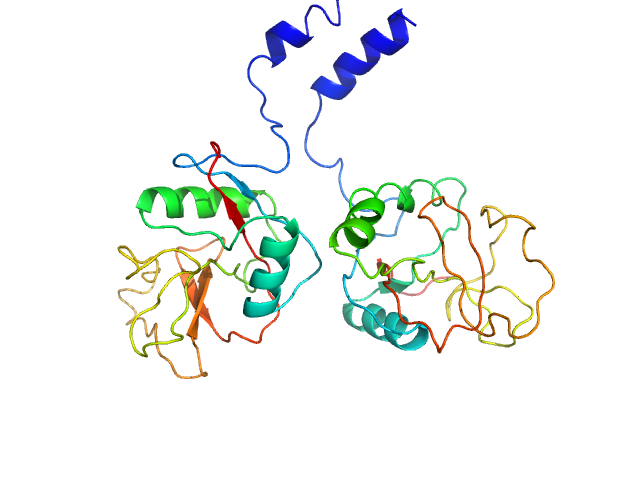
| Sample: |
Aryl-hydrocarbon-interacting protein-like 1(1-316) monomer, 37 kDa Homo sapiens protein
|
| Buffer: |
50 mM Tris, 100 mM NaCl, 2.5 % glycerol and 6 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2018 Jul 17
|
Interaction of the tetratricopeptide repeat domain of aryl hydrocarbon receptor-interacting protein-like 1 with the regulatory Pγ subunit of phosphodiesterase 6.
J Biol Chem 294(43):15795-15807 (2019)
Yadav RP, Boyd K, Yu L, Artemyev NO
|
| RgGuinier |
2.6 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
60 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
AGAP005335-PA monomer, 18 kDa Anopheles gambiae protein
AGAP005334-PA monomer, 18 kDa Anopheles gambiae protein
|
| Buffer: |
500 mM NaCl, 20 mM CHES, 0.5 mM CaCl2, 1% glycerol, pH: 9
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, Thomas Jefferson University on 2018 Aug 30
|
Solution structure, glycan specificity and of phenol oxidase inhibitory activity of Anopheles C-type lectins CTL4 and CTLMA2.
Sci Rep 9(1):15191 (2019)
Bishnoi R, Sousa GL, Contet A, Day CJ, Hou CD, Profitt LA, Singla D, Jennings MP, Valentine AM, Povelones M, Baxter RHG
|
| RgGuinier |
2.5 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
12 base-paired RNA double helix monomer, 8 kDa RNA
|
| Buffer: |
30 mM KCl, 20 mM KMOPS, 20 µM EDTA, pH: 7
|
| Experiment: |
SAXS
data collected at G1, Cornell High Energy Synchrotron Source (CHESS) on 2017 Apr 16
|
Salt Dependence of A-Form RNA Duplexes: Structures and Implications.
J Phys Chem B 123(46):9773-9785 (2019)
Chen YL, Pollack L
|
|
|