|
|
|
|

|
| Sample: |
Zinc protease PqqL monomer, 102 kDa Escherichia coli protein
|
| Buffer: |
20 mM Tris HCl, 150 nM NaCl, 0.02 % NaN3, 5% glycerol, pH: 7.8
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2017 Jun 14
|
Protease-associated import systems are widespread in Gram-negative bacteria.
PLoS Genet 15(10):e1008435 (2019)
Grinter R, Leung PM, Wijeyewickrema LC, Littler D, Beckham S, Pike RN, Walker D, Greening C, Lithgow T
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.7 |
nm |
| VolumePorod |
207 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase tetramer, 131 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.4 |
nm |
| Dmax |
9.0 |
nm |
| VolumePorod |
188 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase (N84D mutant) dimer, 65 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.1 |
nm |
| Dmax |
9.5 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
4-hydroxy-tetrahydrodipicolinate synthase (N84A mutant) , 33 kDa Campylobacter jejuni protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2017 Aug 2
|
Asparagine-84, a regulatory allosteric site residue, helps maintain the quaternary structure of Campylobacter jejuni dihydrodipicolinate synthase.
J Struct Biol :107409 (2019)
Majdi Yazdi M, Saran S, Mrozowich T, Lehnert C, Patel TR, Sanders DAR, Palmer DRJ
|
| RgGuinier |
3.3 |
nm |
| Dmax |
8.9 |
nm |
| VolumePorod |
163 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Alkaline phosphodiesterase I or Nucleotide pyrophosphatase monomer, 46 kDa Streptomyces venezuelae protein
|
| Buffer: |
20 mM HEPES, 200mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 17
|
c-di-AMP hydrolysis by a novel type of phosphodiesterase promotes differentiation of multicellular bacteria
(2019)
Latoscha A, Drexler D, Al-Bassam M, Kaever V, Findlay K, Witte G, Tschowri N
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.8 |
nm |
| VolumePorod |
74 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Cyclic di-AMP binding protein (Putative regulatory, ligand-binding protein) dimer, 38 kDa Streptomyces venezuelae protein
|
| Buffer: |
200 mM NaCl, 30 mM NaPi, 5% (v/v) glycerol, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2019 Nov 4
|
c-di-AMP hydrolysis by a novel type of phosphodiesterase promotes differentiation of multicellular bacteria
(2019)
Latoscha A, Drexler D, Al-Bassam M, Kaever V, Findlay K, Witte G, Tschowri N
|
| RgGuinier |
2.8 |
nm |
| Dmax |
9.2 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
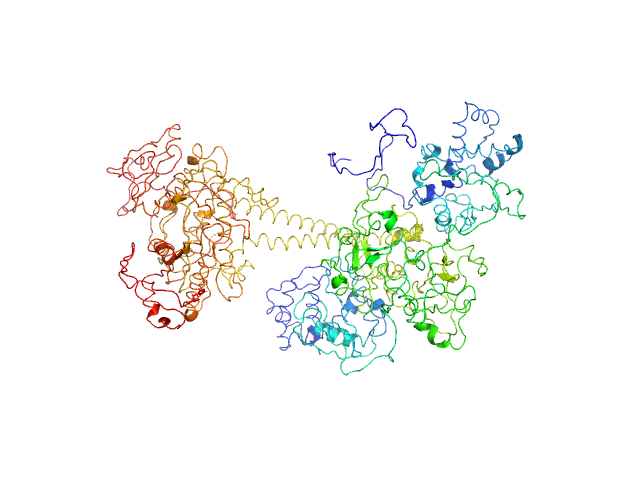
| Sample: |
Soluble guanylyl cyclase alpha-1 subunit monomer, 78 kDa Manduca sexta protein
Soluble guanylyl cyclase beta-1 subunit monomer, 68 kDa Manduca sexta protein
|
| Buffer: |
50 mM KH2PO4, 150 mM NaCl, 2% glycerol,, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 3
|
Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife 8 (2019)
Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, Marletta MA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
13.3 |
nm |
| VolumePorod |
230 |
nm3 |
|
|
|
|
|
|
|
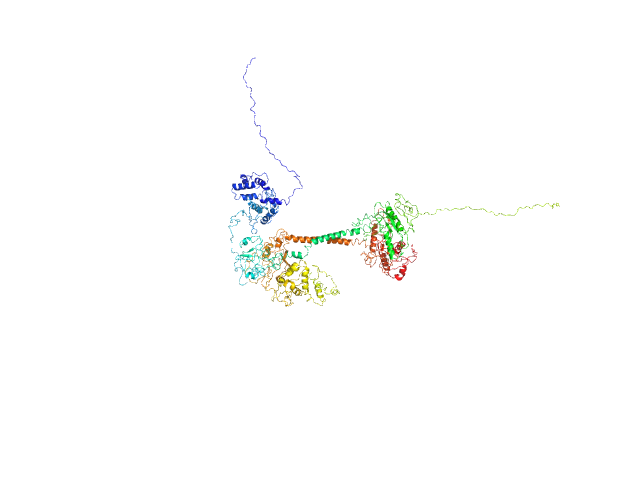
| Sample: |
Soluble guanylyl cyclase alpha-1 subunit monomer, 78 kDa Manduca sexta protein
Soluble guanylyl cyclase beta-1 subunit monomer, 68 kDa Manduca sexta protein
|
| Buffer: |
50 mM KH2PO4,150 mM NaCl, 2% glycerol, 500 µM NO,, pH: 7.4
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Dec 3
|
Allosteric activation of the nitric oxide receptor soluble guanylate cyclase mapped by cryo-electron microscopy.
Elife 8 (2019)
Horst BG, Yokom AL, Rosenberg DJ, Morris KL, Hammel M, Hurley JH, Marletta MA
|
| RgGuinier |
4.4 |
nm |
| Dmax |
14.2 |
nm |
| VolumePorod |
218 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 10 mM NaCl, 0.05% NaN3, pH: 6.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zi...
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
38 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
DNA-directed RNA polymerase subunit delta monomer, 20 kDa Bacillus subtilis protein
|
| Buffer: |
20 mM Phosphate buffer, 200 mM NaCl, 0.05% NaN3, pH: 6.6
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Oct 3
|
Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase.
J Am Chem Soc (2019)
Kuban V, Srb P, Stegnerova H, Padrta P, Zachrdla M, Jasenakova Z, Šanderová H, Vítovská D, Krasny L, Koval T, Dohnalek J, Ziemska-Legi Cka J, Grynberg M, Jarnot P, Gruca A, Jensen MR, Blackledge M, Zi...
|
| RgGuinier |
3.9 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
56 |
nm3 |
|
|