|
|
|
|
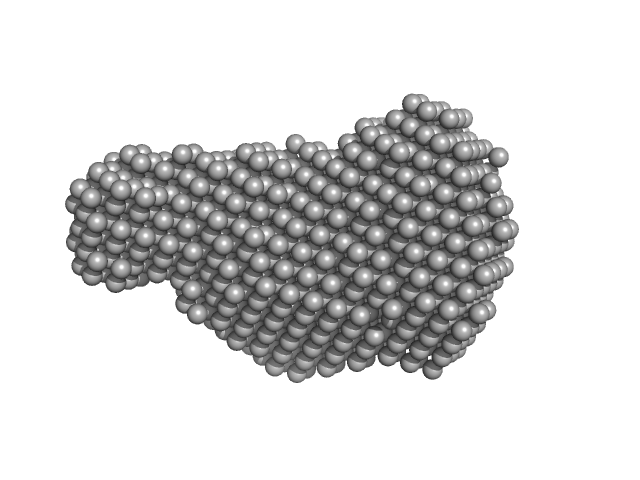
|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
6.8 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Virus termination factor small subunit monomer, 33 kDa Monkeypox virus (strain … protein
mRNA-capping enzyme catalytic subunit monomer, 35 kDa Monkeypox virus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2024 Jan 4
|
Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect 13(1):2369193 (2024)
Chen A, Fang N, Zhang Z, Wen Y, Shen Y, Zhang Y, Zhang L, Zhao G, Ding J, Li J
|
| RgGuinier |
2.9 |
nm |
| Dmax |
10.1 |
nm |
| VolumePorod |
106 |
nm3 |
|
|
|
|
|
|
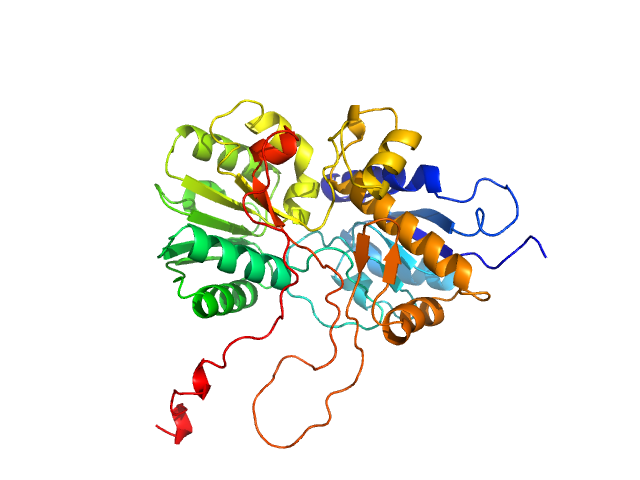
|
| Sample: |
adenylate cyclase monomer, 43 kDa Trypanosoma brucei protein
|
| Buffer: |
50 mM Tris-HCl, 500 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2023 Sep 23
|
Biophysical analysis of the membrane-proximal Venus Flytrap domain of ESAG4 receptor-like adenylate cyclase from Trypanosoma brucei.
Mol Biochem Parasitol 260:111653 (2024)
Alves DO, Geens R, da Silva Arruda HR, Jennen L, Corthaut S, Wuyts E, de Andrade GC, Prosdocimi F, Cordeiro Y, Pires JR, Vieira LR, de Oliveira GAP, Sterckx YG, Salmon D
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
76 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 688-817, i.e. dsRBD3-long) dimer, 29 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.6 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
50 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 708-801, i.e. dsRBD3-mid) dimer, 22 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
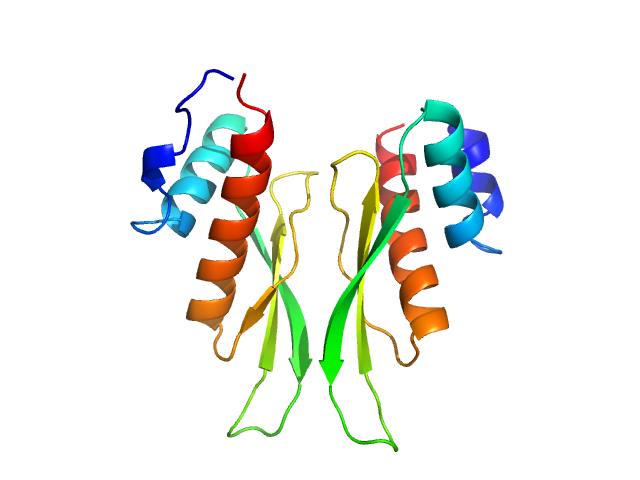
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (residues 716-797, i.e. dsRBD3-short) dimer, 18 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-HEPES, 55 mM KOAc, 10 mM NaCl, 1 mM TCEP, pH: 7.3
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2020 Jun 27
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
1.9 |
nm |
| Dmax |
5.5 |
nm |
| VolumePorod |
27 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (wild-type, residues 708-801, i.e. ADAR1-dsRBD3) dimer, 25 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.1 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Third double-stranded RNA-binding domain of human ADAR1 (interface mutant, residues 708-801, i.e. ADAR1-dsRBD3 interface mutant) monomer, 12 kDa Homo sapiens protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
20 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Chimeric construct between the third dsRBD of human ADAR1 and the second dsRBD of Xenopus Xlrbpa (chimeric construct, residues 708-801, i.e. Chimeric ADAR1-ds3/Xlrbpa-ds2) monomer, 13 kDa Homo sapiens / … protein
|
| Buffer: |
20 mM Na-phosphate, 100 mM NaCl, 2 mM 2-mercaptoethanol, pH: 7
|
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2021 Oct 8
|
Dimerization of ADAR1 modulates site-specificity of RNA editing
Nature Communications 15(1) (2024)
Mboukou A, Rajendra V, Messmer S, Mandl T, Catala M, Tisné C, Jantsch M, Barraud P
|
| RgGuinier |
2.0 |
nm |
| Dmax |
6.4 |
nm |
| VolumePorod |
16 |
nm3 |
|
|
|
|
|
|
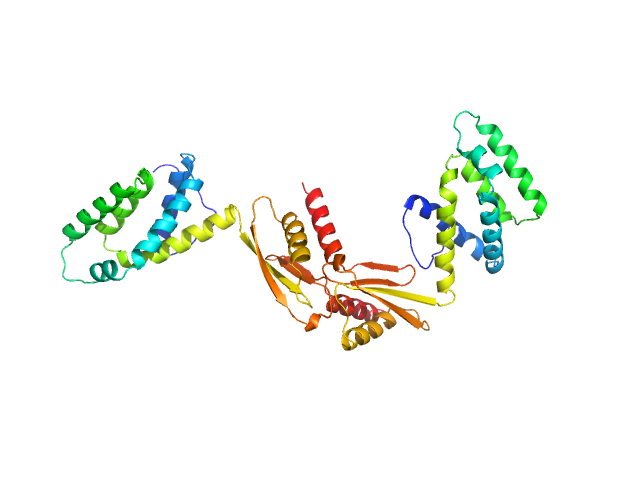
|
| Sample: |
ATP synthase subunit O, mitochondrial dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
50 mM Na2HPO4, 300 mM NaCl, pH: 7
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2022 Mar 15
|
Unveiling the structural properties of the ATP Synthase subunit OSCP by SAXS
Gabriele Giachin
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
78 |
nm3 |
|
|