|
|
|
|
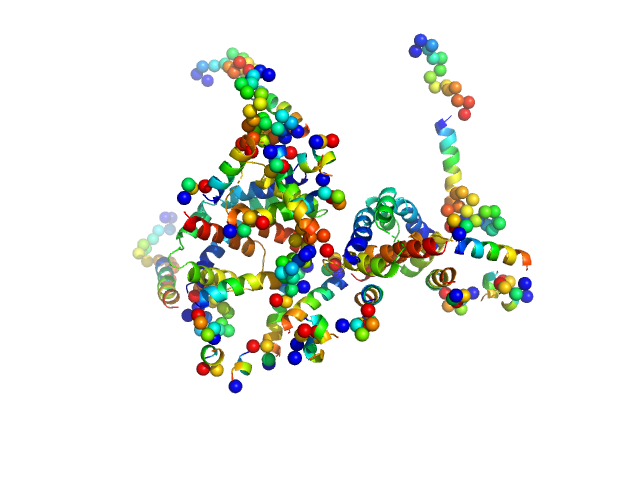
|
| Sample: |
Apoptosis regulator BAX (Bcl-2 associated X) tetramer, 85 kDa Homo sapiens protein
|
| Buffer: |
20mM sodium phosphate 100mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at 23A, Taiwan Photon Source, NSRRC on 2015 May 28
|
Oligomerization process of Bcl-2 associated X protein revealed from intermediate structures in solution.
Phys Chem Chem Phys 19(11):7947-7954 (2017)
Shih O, Yeh YQ, Liao KF, Sung TC, Chiang YW, Jeng US
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
180 |
nm3 |
|
|
|
|
|
|
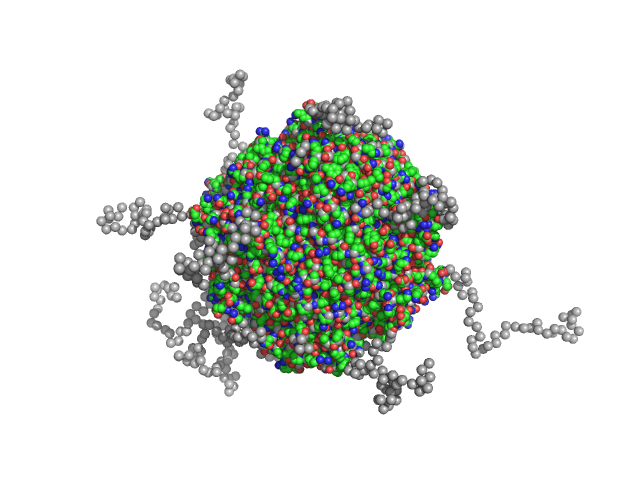
|
| Sample: |
N-terminal truncated DNA protection during starvation protein 2 dodecamer, 279 kDa Deinococcus radiodurans R1 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
SAXS Structural Studies of Dps from Deinococcus radiodurans Highlights the Conformation of the Mobile N-Terminal Extensions.
J Mol Biol 429(5):667-687 (2017)
Santos SP, Cuypers MG, Round A, Finet S, Narayanan T, Mitchell EP, Romão CV
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
445 |
nm3 |
|
|
|
|
|
|
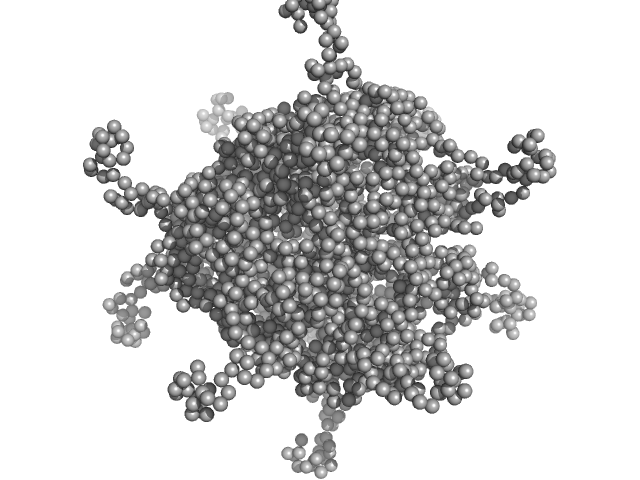
|
| Sample: |
DNA protection during starvation protein 1 dodecamer, 276 kDa Deinococcus radiodurans R1 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
SAXS Structural Studies of Dps from Deinococcus radiodurans Highlights the Conformation of the Mobile N-Terminal Extensions.
J Mol Biol 429(5):667-687 (2017)
Santos SP, Cuypers MG, Round A, Finet S, Narayanan T, Mitchell EP, Romão CV
|
| RgGuinier |
4.2 |
nm |
| Dmax |
12.8 |
nm |
| VolumePorod |
437 |
nm3 |
|
|
|
|
|
|
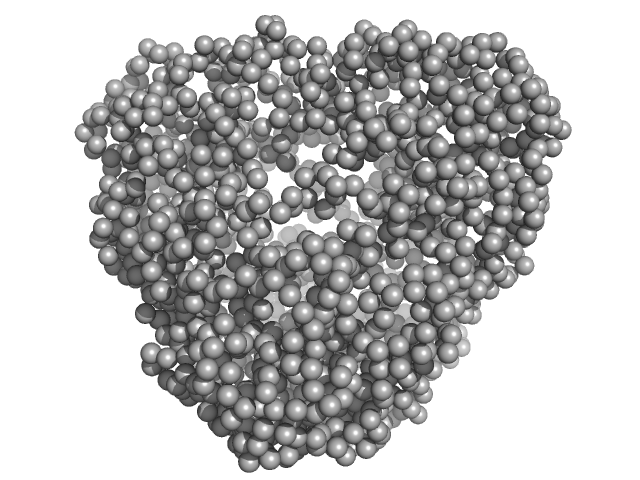
|
| Sample: |
N-terminal truncated DNA protection during starvation protein 1 dodecamer, 216 kDa Deinococcus radiodurans R1 protein
|
| Buffer: |
20 mM Tris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2013 Nov 22
|
SAXS Structural Studies of Dps from Deinococcus radiodurans Highlights the Conformation of the Mobile N-Terminal Extensions.
J Mol Biol 429(5):667-687 (2017)
Santos SP, Cuypers MG, Round A, Finet S, Narayanan T, Mitchell EP, Romão CV
|
| RgGuinier |
3.9 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
291 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-crystallin B2 dimer, 46 kDa Homo sapiens protein
|
| Buffer: |
25 mM NaPi, 5 mM DTT, 1 mM EDTA,, pH: 6.5
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 20
|
Human βB2-Crystallin Forms a Face-en-Face Dimer in Solution: An Integrated NMR and SAXS Study.
Structure 25(3):496-505 (2017)
Xi Z, Whitley MJ, Gronenborn AM
|
| RgGuinier |
2.2 |
nm |
| Dmax |
8.1 |
nm |
| VolumePorod |
67 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Beta-crystallin B2 dimer, 42 kDa Homo sapiens protein
|
| Buffer: |
25 mM NaPi, 5 mM DTT, 1 mM EDTA,, pH: 6.5
|
| Experiment: |
SAXS
data collected at 12-ID-B SAXS/WAXS, Advanced Photon Source (APS), Argonne National Laboratory on 2015 Mar 20
|
Human βB2-Crystallin Forms a Face-en-Face Dimer in Solution: An Integrated NMR and SAXS Study.
Structure 25(3):496-505 (2017)
Xi Z, Whitley MJ, Gronenborn AM
|
| RgGuinier |
2.1 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
62 |
nm3 |
|
|
|
|
|
|
|
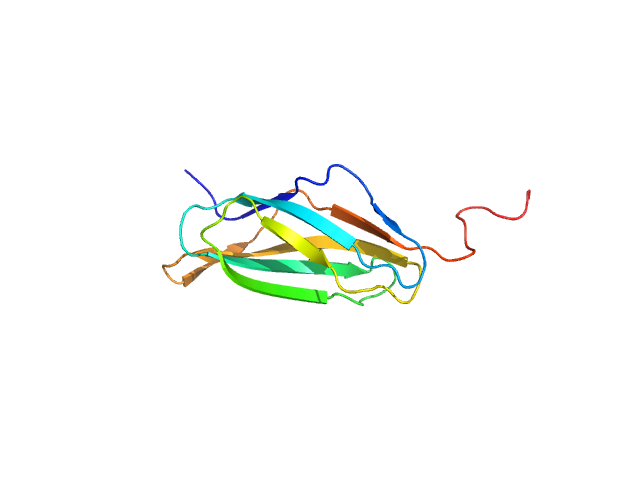
| Sample: |
Nucleoporin POM152 monomer, 12 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 10%(v/v) glycerol, 5mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Apr 12
|
Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure 25(3):434-445 (2017)
Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J
|
| RgGuinier |
1.8 |
nm |
| Dmax |
6.7 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|

|
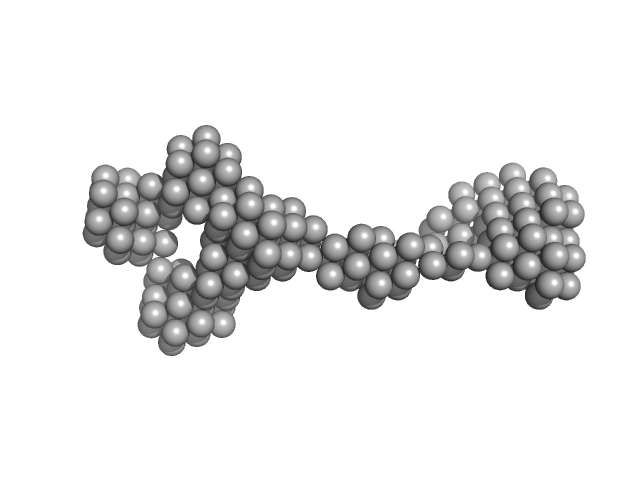
| Sample: |
Nucleoporin POM152 monomer, 24 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 10%(v/v) glycerol, 5mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Apr 12
|
Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure 25(3):434-445 (2017)
Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J
|
| RgGuinier |
2.7 |
nm |
| Dmax |
9.4 |
nm |
| VolumePorod |
23 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Nucleoporin POM152 monomer, 12 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 10%(v/v) glycerol, 5mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Apr 12
|
Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure 25(3):434-445 (2017)
Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J
|
| RgGuinier |
2.8 |
nm |
| Dmax |
7.9 |
nm |
| VolumePorod |
57 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Nucleoporin POM152 monomer, 26 kDa Saccharomyces cerevisiae protein
|
| Buffer: |
10mM HEPES, 150mM NaCl, 10%(v/v) glycerol, 5mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at BL4-2, Stanford Synchrotron Radiation Lightsource (SSRL) on 2015 Apr 12
|
Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex.
Structure 25(3):434-445 (2017)
Upla P, Kim SJ, Sampathkumar P, Dutta K, Cahill SM, Chemmama IE, Williams R, Bonanno JB, Rice WJ, Stokes DL, Cowburn D, Almo SC, Sali A, Rout MP, Fernandez-Martinez J
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.5 |
nm |
| VolumePorod |
28 |
nm3 |
|
|