|
|
|
|
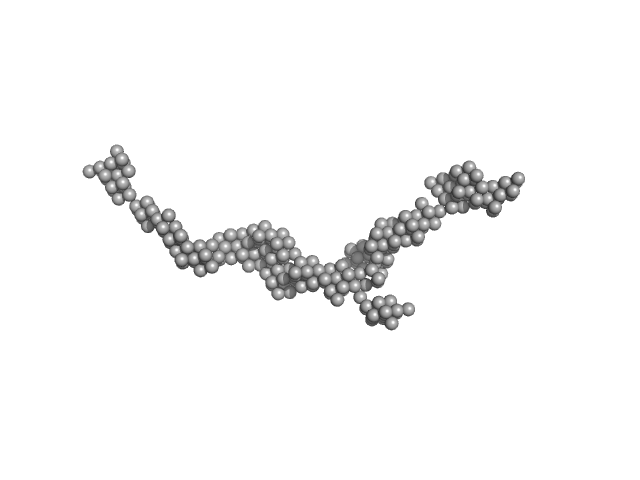
|
| Sample: |
Myomesin-1 dimer, 121 kDa Homo sapiens protein
|
| Buffer: |
25 mMTris-HCl, 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 May 10
|
Superhelical architecture of the myosin filament-linking protein myomesin with unusual elastic properties.
PLoS Biol 10(2):e1001261 (2012)
Pinotsis N, Chatziefthimiou SD, Berkemeier F, Beuron F, Mavridis IM, Konarev PV, Svergun DI, Morris E, Rief M, Wilmanns M
|
| RgGuinier |
9.6 |
nm |
| Dmax |
37.0 |
nm |
|
|
|
|
|
|
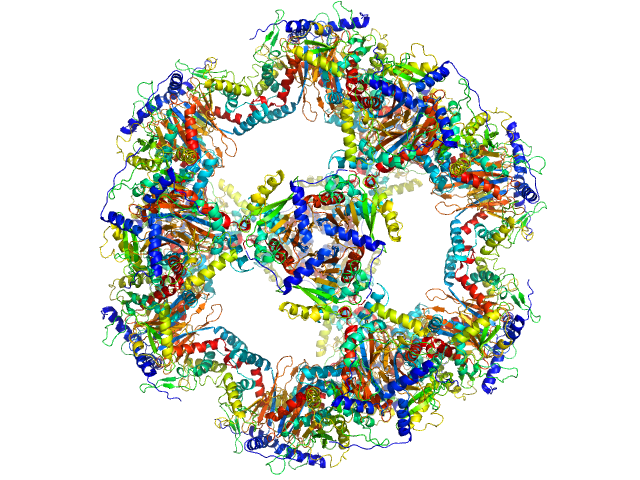
|
| Sample: |
Regulatory protein E2 , 1037 kDa Human papillomavirus type … protein
|
| Buffer: |
50 mM Tris ⁄ HCl, pH 8.8, 100 mM NaCl, pH: 8.8
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Jul 13
|
The catalytic core of an archaeal 2-oxoacid dehydrogenase multienzyme complex is a 42-mer protein assembly
FEBS Journal 279(5):713-723 (2012)
Marrott N, Marshall J, Svergun D, Crennell S, Hough D, Danson M, van den Elsen J
|
| RgGuinier |
8.8 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
2473 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
Full length GbpA monomer, 54 kDa Vibrio cholerae protein
|
| Buffer: |
25 mM Tris/HCl 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 16
|
The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces.
PLoS Pathog 8(1):e1002373 (2012)
Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AF, Svergun DI, Eijsink VG, Chatterjee NS, van Aalten DM
|
| RgGuinier |
3.9 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
100 |
nm3 |
|
|
|
|
|
|
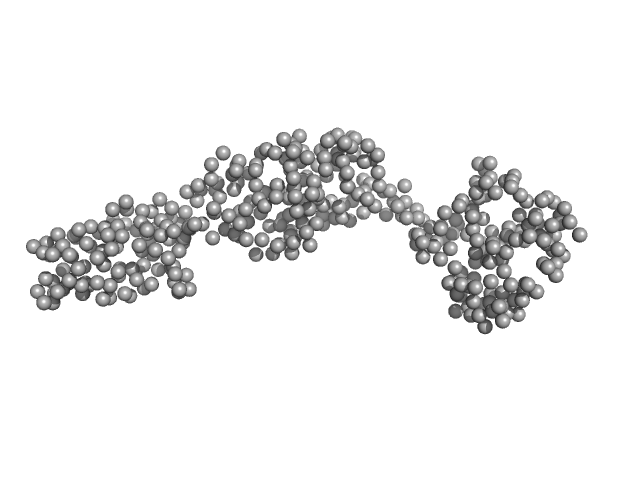
|
| Sample: |
Truncated GbpA monomer, 44 kDa Vibrio cholerae protein
|
| Buffer: |
25 mM Tris/HCl 150 mM NaCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Oct 16
|
The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces.
PLoS Pathog 8(1):e1002373 (2012)
Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AF, Svergun DI, Eijsink VG, Chatterjee NS, van Aalten DM
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.5 |
nm |
| VolumePorod |
80 |
nm3 |
|
|
|
|
|
|
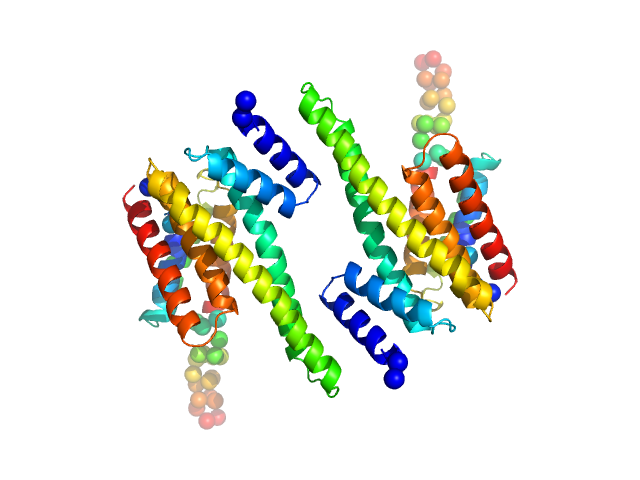
|
| Sample: |
14-3-3 protein beta/alpha dimer, 58 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.0 |
nm |
| Dmax |
10.0 |
nm |
| VolumePorod |
92 |
nm3 |
|
|
|
|
|
|
|
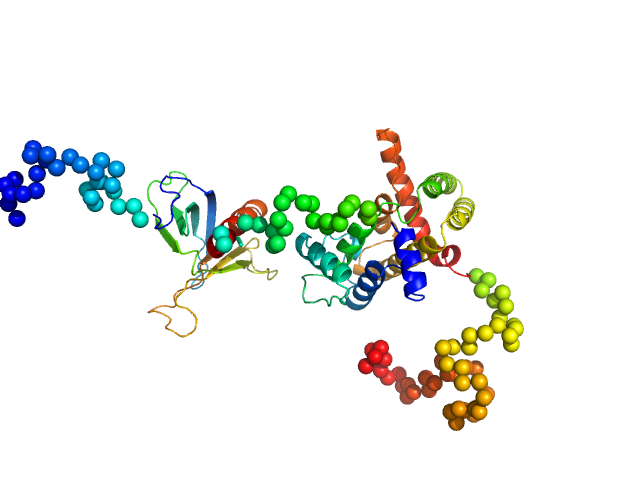
| Sample: |
Rho GTPase-activating protein 22 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.2 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
88 |
nm3 |
|
|
|
|
|
|
|
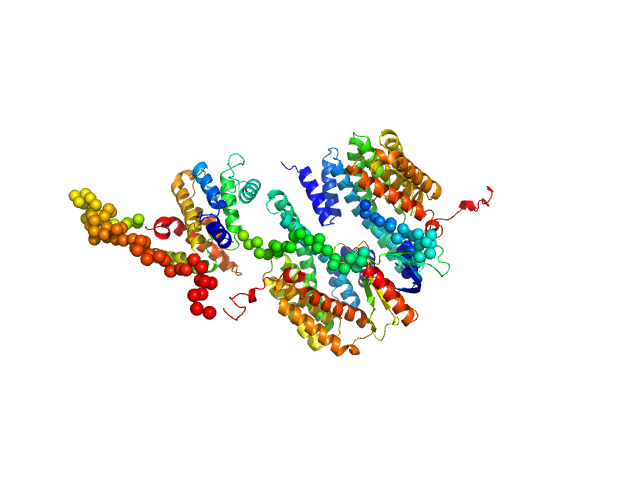
| Sample: |
14-3-3 protein beta/alpha dimer, 58 kDa Homo sapiens protein
Rho GTPase-activating protein 22 monomer, 47 kDa Homo sapiens protein
|
| Buffer: |
25 mM HEPES, 150 mM NaCl, and 2 mM 2-mercaptoethanol, pH: 7.5
|
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2011 Apr 7
|
The weak complex between RhoGAP protein ARHGAP22 and signal regulatory protein 14-3-3 has 1:2 stoichiometry and a single peptide binding mode.
PLoS One 7(8):e41731 (2012)
Hu SH, Whitten AE, King GJ, Jones A, Rowland AF, James DE, Martin JL
|
| RgGuinier |
3.8 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
195 |
nm3 |
|
|
|
|
|
|
|
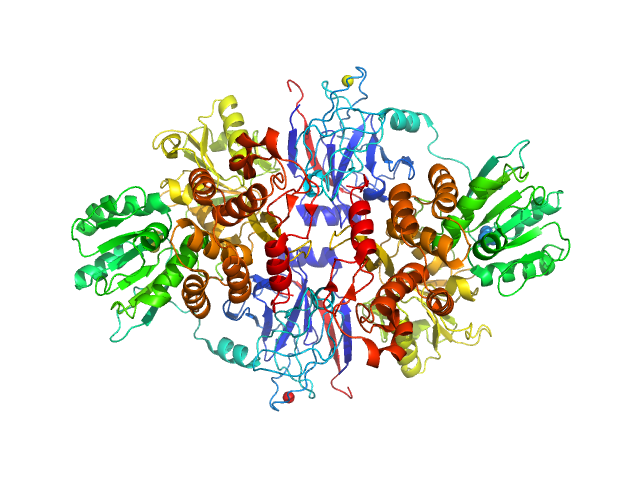
| Sample: |
Genome polyprotein dimer, 137 kDa Hepatitis C virus … protein
|
| Buffer: |
25 mM Tris, 1 M NaCl, 10% glycerol, 1 mM TCEP, 0.1% β-octyl glucoside, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2007 Dec 12
|
A macrocyclic HCV NS3/4A protease inhibitor interacts with protease and helicase residues in the complex with its full-length target
Proceedings of the National Academy of Sciences 108(52):21052-21056 (2011)
Schiering N, D'Arcy A, Villard F, Simic O, Kamke M, Monnet G, Hassiepen U, Svergun D, Pulfer R, Eder J, Raman P, Bodendorf U
|
|
|
|
|
|
|
|
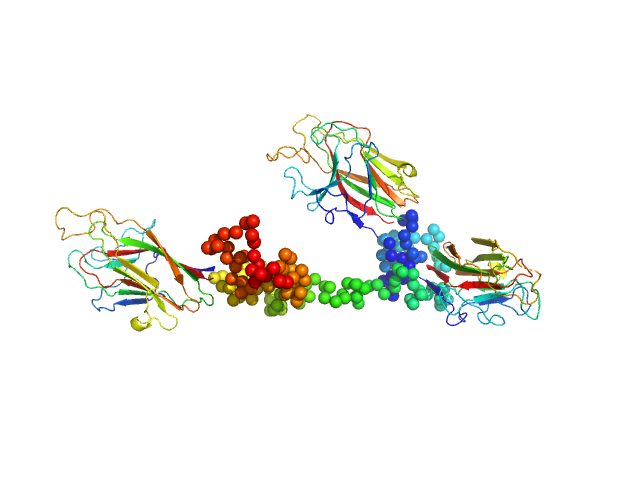
| Sample: |
K1K2K3 adhesin modules of lysine-specific (Kgp) gingipain monomer, 74 kDa Porphyromonas gingivalis W83 protein
|
| Buffer: |
10 mM TRIS 150 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Sydney on 2010 Aug 15
|
The modular structure of haemagglutinin/adhesin regions in gingipains of Porphyromonas gingivalis.
Mol Microbiol 81(5):1358-73 (2011)
Li N, Yun P, Jeffries CM, Langley D, Gamsjaeger R, Church WB, Hunter N, Collyer CA
|
| RgGuinier |
4.3 |
nm |
| Dmax |
14.7 |
nm |
|
|
|
|
|
|
|
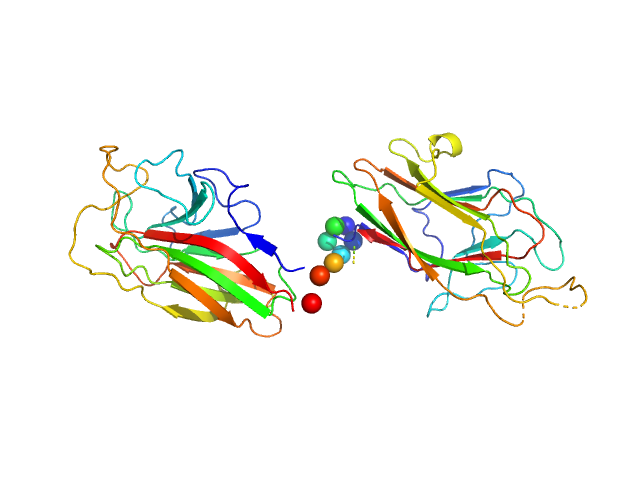
| Sample: |
K1K2 adhesin modules of lysine-specific (Kgp) gingipain monomer, 38 kDa Porphyromonas gingivalis W83 protein
|
| Buffer: |
10 mM TRIS 150 mM NaCl, pH: 7.6
|
| Experiment: |
SAXS
data collected at Anton Paar SAXSess, University of Sydney on 2010 Aug 15
|
The modular structure of haemagglutinin/adhesin regions in gingipains of Porphyromonas gingivalis.
Mol Microbiol 81(5):1358-73 (2011)
Li N, Yun P, Jeffries CM, Langley D, Gamsjaeger R, Church WB, Hunter N, Collyer CA
|
| RgGuinier |
2.9 |
nm |
| Dmax |
9.5 |
nm |
|
|