|
|
|
|

|
| Sample: |
CYNEX4 FRET probe, (eYFP-AnnexinA4-eCFP) T266D mutant monomer, 93 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
146 |
nm3 |
|
|
|
|
|
|
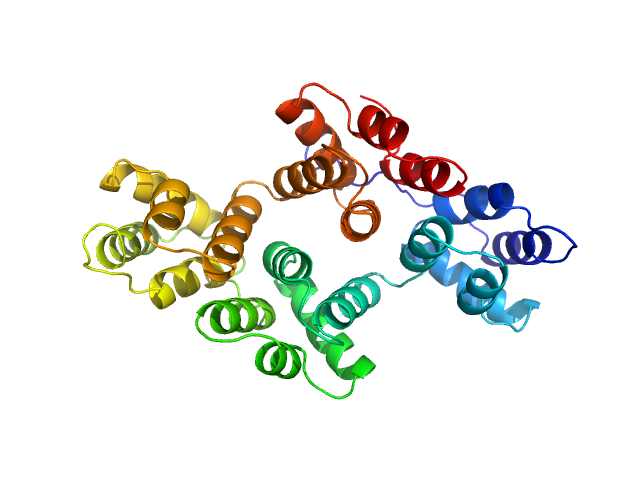
|
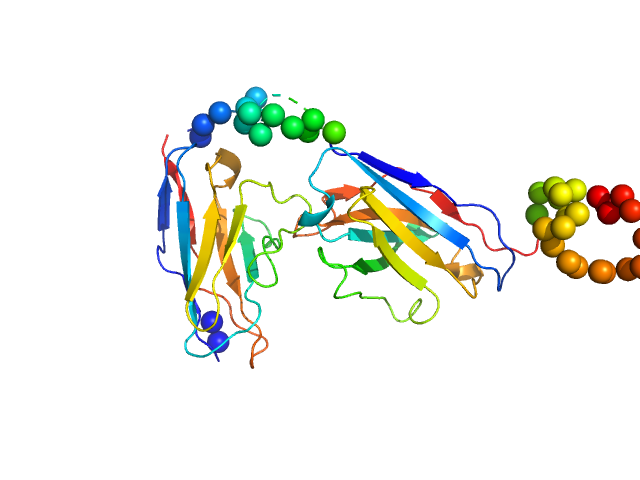
| Sample: |
Annexin-A4 monomer, 36 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES 50 mM KCl, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2010 Sep 11
|
Conformational analysis of a genetically encoded FRET biosensor by SAXS.
Biophys J 102(12):2866-75 (2012)
Mertens HD, Piljić A, Schultz C, Svergun DI
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.6 |
nm |
| VolumePorod |
54 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Recombinant monoclonal anti-proNGF antibody in single chain Fv fragment (scFv) monomer, 29 kDa protein
|
| Buffer: |
phosphate buffered saline, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2005 Oct 21
|
Direct intracellular selection and biochemical characterization of a recombinant anti-proNGF single chain antibody fragment.
Arch Biochem Biophys 522(1):26-36 (2012)
Paoletti F, Malerba F, Konarev PV, Visintin M, Scardigli R, Fasulo L, Lamba D, Svergun DI, Cattaneo A
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
39 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Frataxin homolog, mitochondrial monomer, 14 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM HEPES, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 28
|
The role of hydration in protein stability: comparison of the cold and heat unfolded states of Yfh1.
J Mol Biol 417(5):413-24 (2012)
Adrover M, Martorell G, Martin SR, Urosev D, Konarev PV, Svergun DI, Daura X, Temussi P, Pastore A
|
|
|
|
|
|
|
|
| Sample: |
Frataxin homolog, mitochondrial monomer, 14 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM HEPES, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 28
|
The role of hydration in protein stability: comparison of the cold and heat unfolded states of Yfh1.
J Mol Biol 417(5):413-24 (2012)
Adrover M, Martorell G, Martin SR, Urosev D, Konarev PV, Svergun DI, Daura X, Temussi P, Pastore A
|
|
|
|
|
|
|
|
| Sample: |
Frataxin homolog, mitochondrial monomer, 14 kDa Saccharomyces cerevisiae (strain … protein
|
| Buffer: |
20 mM HEPES, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2008 Nov 28
|
The role of hydration in protein stability: comparison of the cold and heat unfolded states of Yfh1.
J Mol Biol 417(5):413-24 (2012)
Adrover M, Martorell G, Martin SR, Urosev D, Konarev PV, Svergun DI, Daura X, Temussi P, Pastore A
|
|
|
|
|
|
|
|
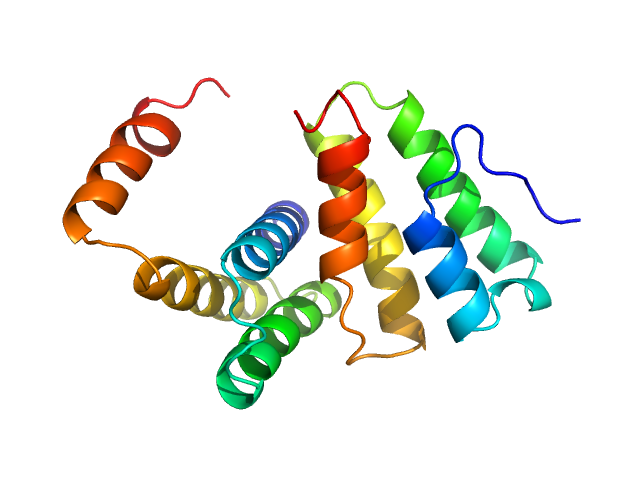
| Sample: |
Polyglutamine-binding protein 1 monomer, 31 kDa Homo sapiens protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 1mM DTT,, pH: 7
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2009 Nov 18
|
Solution model of the intrinsically disordered polyglutamine tract-binding protein-1.
Biophys J 102(7):1608-16 (2012)
Rees M, Gorba C, de Chiara C, Bui TT, Garcia-Maya M, Drake AF, Okazawa H, Pastore A, Svergun D, Chen YW
|
| RgGuinier |
3.7 |
nm |
| Dmax |
13.0 |
nm |
| VolumePorod |
51 |
nm3 |
|
|
|
|
|
|
|
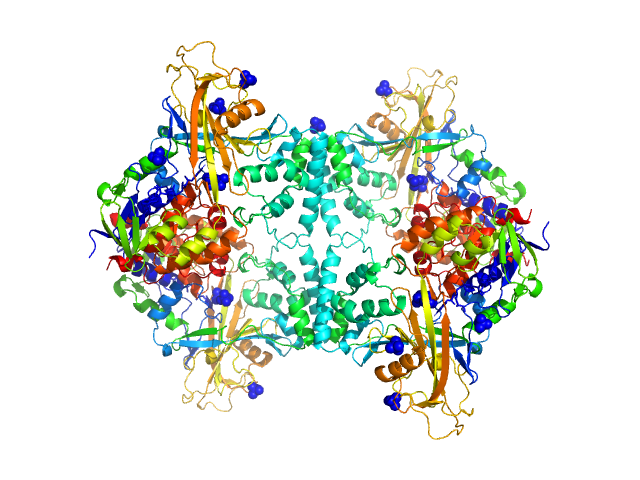
| Sample: |
Aspergillus fumigatus UDP galactopyranose mutase tetramer, 228 kDa protein
|
| Buffer: |
20 mM HEPES, 45 mM NaCl, 0.5 mM Tris(hydroxypropyl)phosphine, pH: 7.5
|
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2010 Apr 19
|
Crystal structures and small-angle x-ray scattering analysis of UDP-galactopyranose mutase from the pathogenic fungus Aspergillus fumigatus.
J Biol Chem 287(12):9041-51 (2012)
Dhatwalia R, Singh H, Oppenheimer M, Karr DB, Nix JC, Sobrado P, Tanner JJ
|
| RgGuinier |
4.7 |
nm |
| Dmax |
14.7 |
nm |
| VolumePorod |
308 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Replicase polyprotein 1a dimer, 19 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Apr 2
|
Nonstructural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer That Exhibits Primer-Independent RNA Polymerase Activity
Journal of Virology 86(8):4444-4454 (2012)
Xiao Y, Ma Q, Restle T, Shang W, Svergun D, Ponnusamy R, Sczakiel G, Hilgenfeld R
|
|
|
|
|
|
|
|
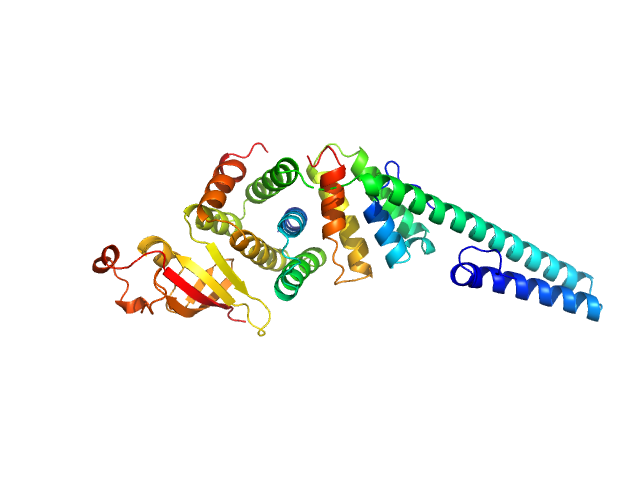
| Sample: |
Replicase polyprotein 1a dimer, 19 kDa Severe acute respiratory … protein
Replicase polyprotein 1a monomer, 22 kDa Severe acute respiratory … protein
|
| Buffer: |
10 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, pH: 7.5
|
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2011 Apr 2
|
Nonstructural Proteins 7 and 8 of Feline Coronavirus Form a 2:1 Heterotrimer That Exhibits Primer-Independent RNA Polymerase Activity
Journal of Virology 86(8):4444-4454 (2012)
Xiao Y, Ma Q, Restle T, Shang W, Svergun D, Ponnusamy R, Sczakiel G, Hilgenfeld R
|
|
|