|
|
|
|
|
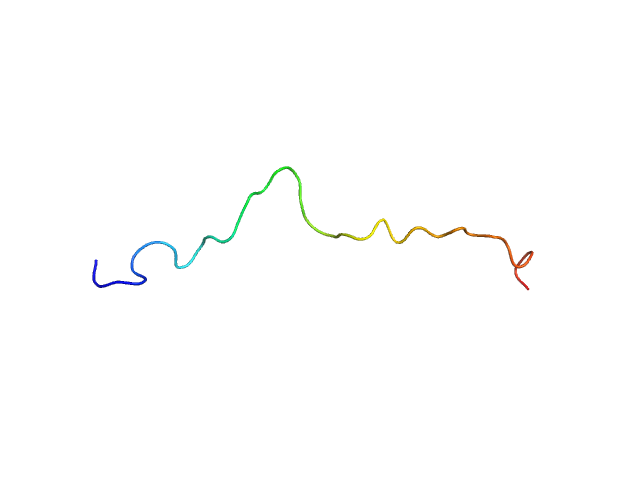
| Sample: |
HTT0 monomer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2023 Nov 10
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
1.5 |
nm |
| Dmax |
6.0 |
nm |
| VolumePorod |
4 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
HTT1 dimer, 8 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Feb 3
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
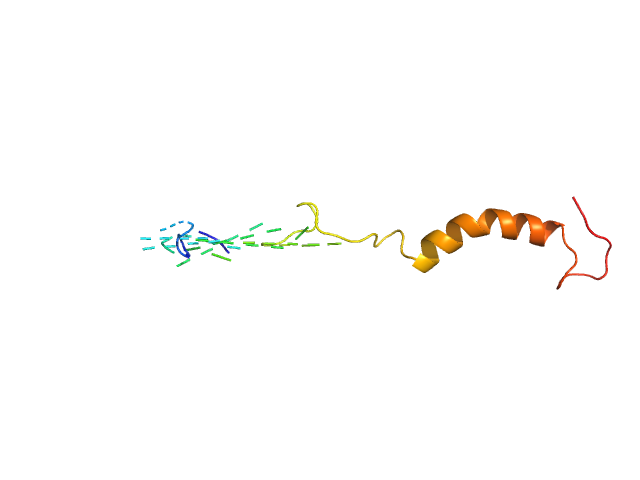
|
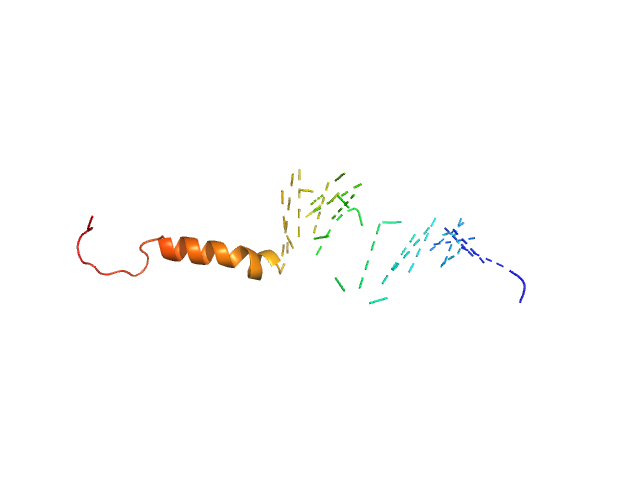
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
NT17 dimer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Oct 21
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
|
|
|
|
|
|
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
HTT3 monomer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 May 20
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA repair protein RAD52 homolog decamer, 234 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-Tris, 10% glycerol, 400 mM NaCl, 100 mM KCl, 1 mM EDTA, pH: 6
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS 1000, Eppley Structural Biology Facility, University of Nebraska Medical Center on 2014 Jul 25
|
A glimpse into the hidden world of the flexible C-terminal protein binding domains of human RAD52
Journal of Structural Biology 216(3):108115 (2024)
Struble L, Lovelace J, Borgstahl G
|
| RgGuinier |
4.1 |
nm |
| Dmax |
11.8 |
nm |
| VolumePorod |
375 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
DNA repair protein RAD52 homolog nonamer, 300 kDa Homo sapiens protein
|
| Buffer: |
20 mM Bis-Tris, 10% GLycerol, 400 mM NaCl, 100 mM KCl, 1mM EDTA, pH: 6
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS 1000, Eppley Structural Biology Facility, University of Nebraska Medical Center on 2014 Jul 25
|
A glimpse into the hidden world of the flexible C-terminal protein binding domains of human RAD52
Journal of Structural Biology 216(3):108115 (2024)
Struble L, Lovelace J, Borgstahl G
|
| RgGuinier |
4.8 |
nm |
| Dmax |
18.3 |
nm |
| VolumePorod |
782 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Isoform A1-A of Heterogeneous nuclear ribonucleoprotein A1 monomer, 13 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 19
|
Design of intrinsically disordered protein variants with diverse structural properties
Science Advances 10(35) (2024)
Pesce F, Bremer A, Tesei G, Hopkins J, Grace C, Mittag T, Lindorff-Larsen K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
V2 variant of the low complexity domain of hnRNPA1 monomer, 13 kDa synthetic construct protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 19
|
Design of intrinsically disordered protein variants with diverse structural properties
Science Advances 10(35) (2024)
Pesce F, Bremer A, Tesei G, Hopkins J, Grace C, Mittag T, Lindorff-Larsen K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
8.3 |
nm |
| VolumePorod |
19 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
V3 variant of the low complexity domain of hnRNPA1 monomer, 13 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 19
|
Design of intrinsically disordered protein variants with diverse structural properties
Science Advances 10(35) (2024)
Pesce F, Bremer A, Tesei G, Hopkins J, Grace C, Mittag T, Lindorff-Larsen K
|
| RgGuinier |
2.3 |
nm |
| Dmax |
9.3 |
nm |
| VolumePorod |
18 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
V4 variant of the low complexity domain of hnRNPA1 monomer, 13 kDa protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 2 mM DTT, pH: 7
|
| Experiment: |
SAXS
data collected at BioCAT 18ID, Advanced Photon Source (APS), Argonne National Laboratory on 2022 Nov 19
|
Design of intrinsically disordered protein variants with diverse structural properties
Science Advances 10(35) (2024)
Pesce F, Bremer A, Tesei G, Hopkins J, Grace C, Mittag T, Lindorff-Larsen K
|
| RgGuinier |
2.4 |
nm |
| Dmax |
9.9 |
nm |
| VolumePorod |
19 |
nm3 |
|
|