|
|
|
|
|
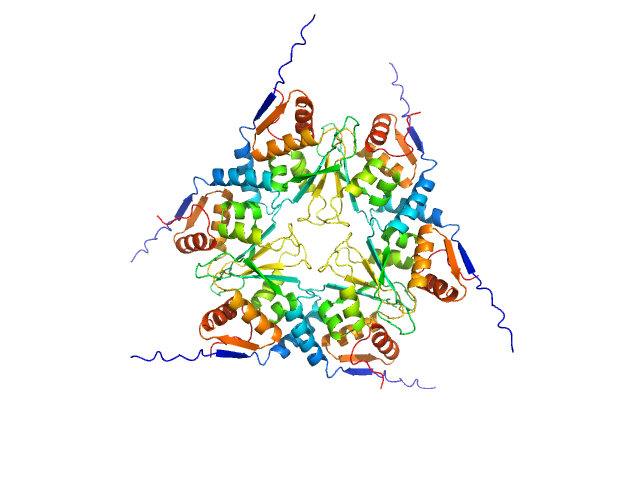
| Sample: |
Glutamate receptor ionotropic, NMDA 1 dimer, 193 kDa Homo sapiens protein
Glutamate receptor ionotropic, NMDA 2A dimer, 191 kDa Homo sapiens protein
Human derived autoantibody mAb5F6 heavy chain, mAb5F6 VH dimer, 104 kDa Homo sapiens protein
Human derived autoantibody mAb5F6 light chain, mAb5F6 VL dimer, 52 kDa protein
|
| Buffer: |
150 mM NaCl, 0.1% digitonin, 5 µM Cholesteryl Hemisuccinate TRIS Salt, 0.1 mM CHAPSO, 50 µM EDTA,1 mM Gly/Glu, 20 mM HEPES, pH: 8
|
| Experiment: |
SAXS
data collected at BL19U2, Shanghai Synchrotron Radiation Facility (SSRF) on 2022 Dec 8
|
Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Nat Struct Mol Biol (2024)
Wang H, Xie C, Deng B, Ding J, Li N, Kou Z, Jin M, He J, Wang Q, Wen H, Zhang J, Zhou Q, Chen S, Chen X, Yuan TF, Zhu S
|
| RgGuinier |
9.9 |
nm |
| Dmax |
31.7 |
nm |
| VolumePorod |
2500 |
nm3 |
|
|
|
|
|
|
|
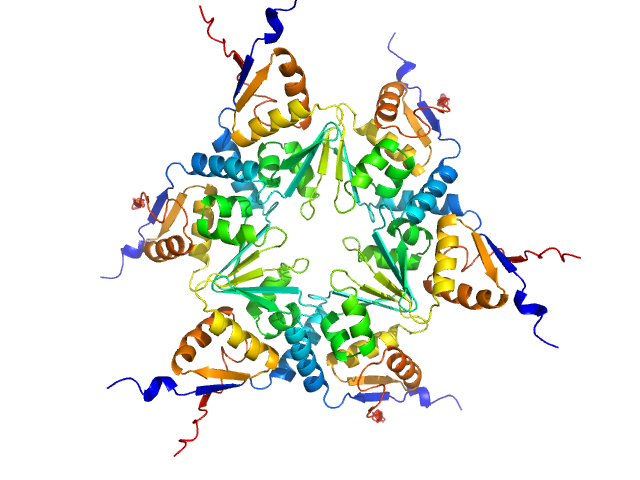
| Sample: |
Uncharacterized protein, isoform A hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 20
|
The Arthropoda-specific Tramtrack group BTB protein domains use previously unknown interface to form hexamers.
Elife 13 (2024)
Bonchuk AN, Balagurov KI, Baradaran R, Boyko KM, Sluchanko NN, Khrustaleva AM, Burtseva AD, Arkova OV, Khalisova KK, Popov VO, Naschberger A, Georgiev PG
|
| RgGuinier |
3.7 |
nm |
| Dmax |
12.7 |
nm |
| VolumePorod |
166 |
nm3 |
|
|
|
|
|
|
|
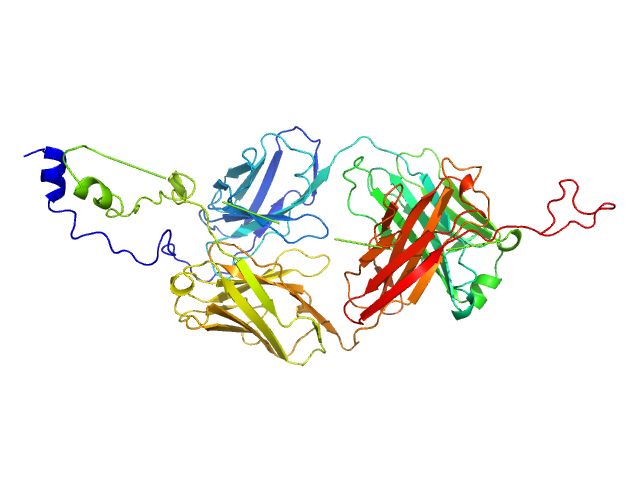
| Sample: |
Longitudinals lacking protein, isoform G hexamer, 92 kDa Drosophila melanogaster protein
|
| Buffer: |
20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM DTT, pH: 7.4
|
| Experiment: |
SAXS
data collected at BM29, ESRF on 2016 Jul 20
|
The Arthropoda-specific Tramtrack group BTB protein domains use previously unknown interface to form hexamers.
Elife 13 (2024)
Bonchuk AN, Balagurov KI, Baradaran R, Boyko KM, Sluchanko NN, Khrustaleva AM, Burtseva AD, Arkova OV, Khalisova KK, Popov VO, Naschberger A, Georgiev PG
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
189 |
nm3 |
|
|
|
|
|
|
|
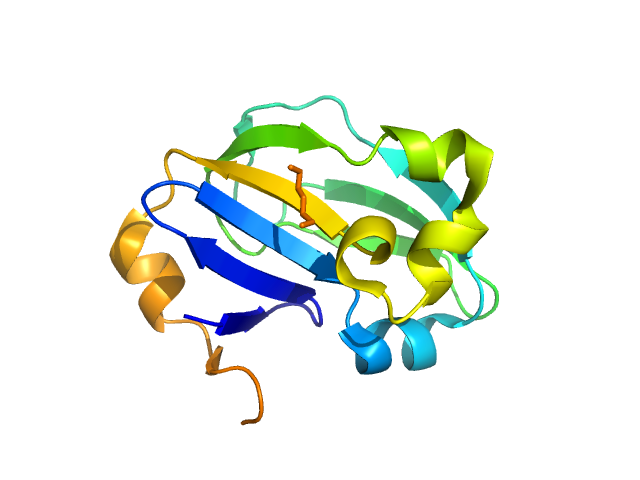
| Sample: |
Pro-Nivolumab, Lu02 monomer, 54 kDa protein
|
| Buffer: |
20 mM Tris, 100 mM NaCl, pH: 8
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2022 Sep 8
|
Integrating molecular dynamics simulation with small- and wide-angle X-ray scattering to unravel the flexibility, antigen-blocking, and protease-restoring functions in a hindrance-based pro-antibody.
Protein Sci 33(9):e5124 (2024)
Liao JM, Hong ST, Wang YT, Cheng YA, Ho KW, Toh SI, Shih O, Jeng US, Lyu PC, Hu IC, Huang MY, Chang CY, Cheng TL
|
| RgGuinier |
3.0 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
69 |
nm3 |
|
|
|
|
|
|
|
| Sample: |
Glycine cleavage system H-like protein monomer, 13 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
22 |
nm3 |
|
|
|
|
|
|
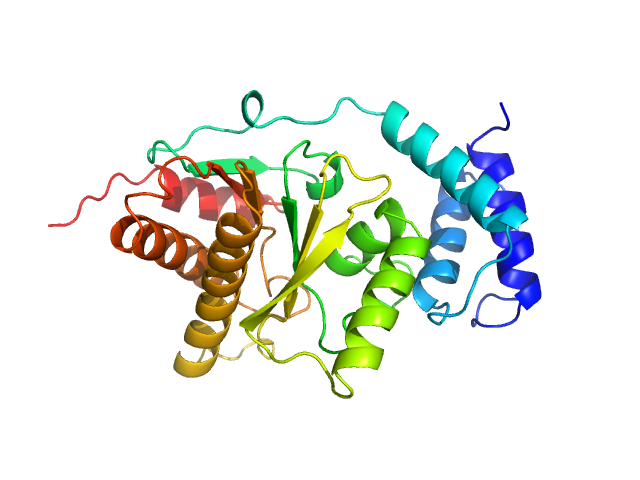
|
| Sample: |
Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T) monomer, 30 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
2.1 |
nm |
| Dmax |
7.3 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
|
|
|
|
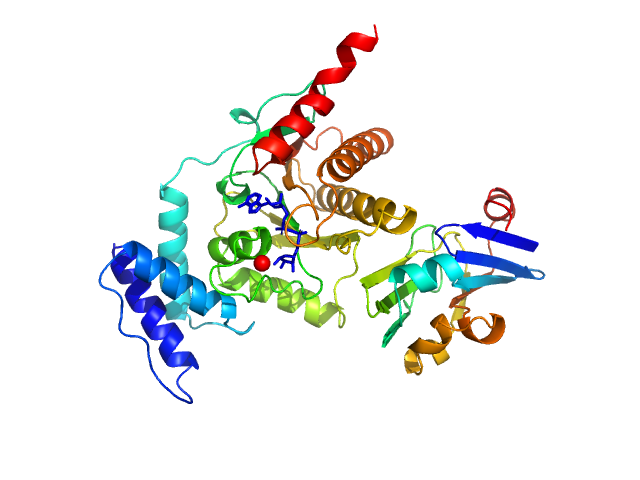
|
| Sample: |
Glycine cleavage system H-like protein monomer, 13 kDa Streptococcus pyogenes serotype … protein
Protein-ADP-ribose hydrolase (D13G, Y23S, T61A, I114S, R177H, I246T) monomer, 30 kDa Streptococcus pyogenes serotype … protein
|
| Buffer: |
50 mM Tris-HCl, 200 mM NaCl, 1 mM DTT, 5% (v/v) glycerol, pH: 8
|
| Experiment: |
SAXS
data collected at B21, Diamond Light Source on 2016 Jul 20
|
Evolutionary and molecular basis of ADP-ribosylation reversal by zinc-dependent macrodomains
Journal of Biological Chemistry :107770 (2024)
Ariza A, Liu Q, Cowieson N, Ahel I, Filippov D, Rack J
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.4 |
nm |
| VolumePorod |
53 |
nm3 |
|
|
|
|
|
|
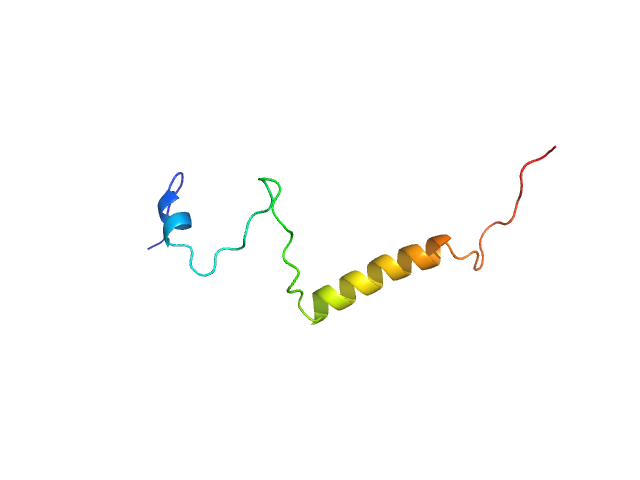
|
| Sample: |
Isoform Short of Small EDRK-rich factor 1 monomer, 7 kDa Homo sapiens protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Mar 11
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
2.4 |
nm |
| Dmax |
8.0 |
nm |
| VolumePorod |
9 |
nm3 |
|
|
|
|
|
|

|
| Sample: |
HTT3 monomer, 4 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 May 20
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
|
|
|
|
|
|

|
| Sample: |
NT17 monomer, 2 kDa synthetic construct protein
|
| Buffer: |
Sodium phosphate buffer, pH: 7.4
|
| Experiment: |
SAXS
data collected at TPS13A, NSRRC on 2021 Oct 21
|
Binding structures of SERF1a with NT17-polyQ peptides of huntingtin exon 1 revealed by SEC-SWAXS, NMR and molecular simulation.
IUCrJ (2024)
Lin TC, Shih O, Tsai TY, Yeh YQ, Liao KF, Mansel BW, Shiu YJ, Chang CF, Su AC, Chen YR, Jeng US
|
| RgGuinier |
1.2 |
nm |
| Dmax |
4.0 |
nm |
| VolumePorod |
5 |
nm3 |
|
|