|
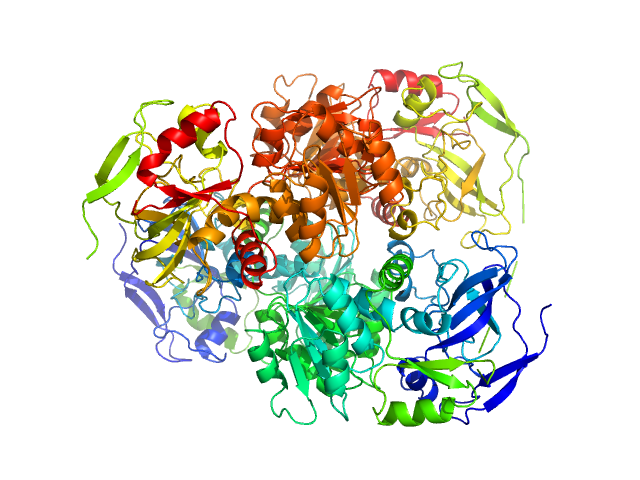
Synchrotron SAXS data from solutions of alcohol dehydrogenase 1 separated using asymmetric flow field flow fractionation (AF4) in phosphate buffered saline, 1% glycerol, pH 7.2 were collected on the EMBL P12 beam line at PETRA III (DESY; Hamburg, Germany) using a Pilatus 6M detector at a sample-detector distance of 3 m and at a wavelength of λ = 0.12 nm (I(s) vs s, where s = 4πsinθ/λ, and 2θ is the scattering angle). AF4 separation was performed at 20°C using a flow rate of 0.5 mL/min with an applied constant cross flow rate of 4 mL/min. 50 µL of alcohol dehydrogenase 1 at a concentration of 7.7 mg/mL were injected. 1500 successive 1 second SAXS data frames were collected through the entire AF4 elution (45 min), where 10 sample data frames were selected through the tetrameric separated elution peak. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank was subtracted.
|
|
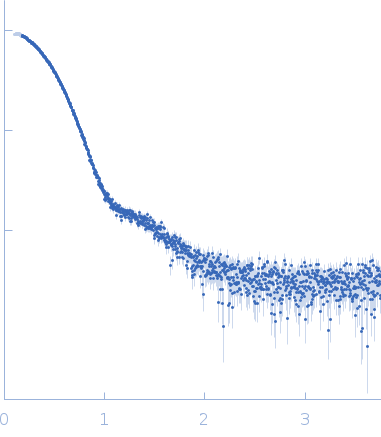 s, nm-1
s, nm-1