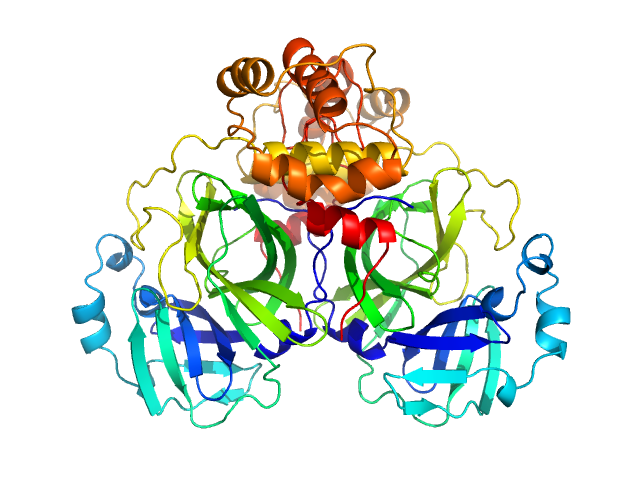
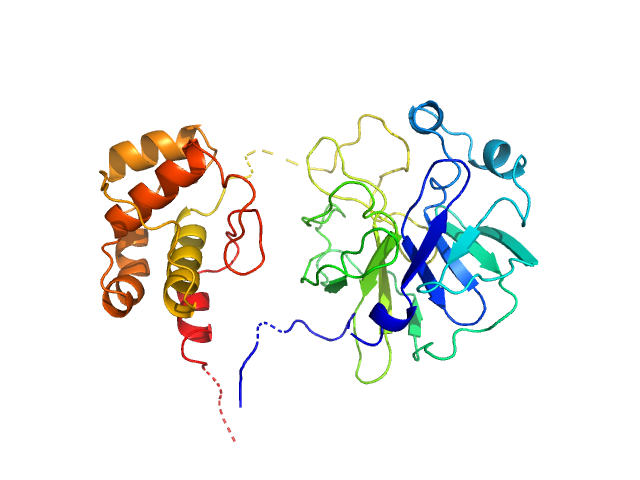
Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Lee J,
Worrall LJ,
Vuckovic M,
Rosell FI,
Gentile F,
Ton AT,
Caveney NA,
Ban F,
Cherkasov A,
Paetzel M,
Strynadka NCJ
Nat Commun
11(1):5877
(2020 Nov 18)
|
|
|
|
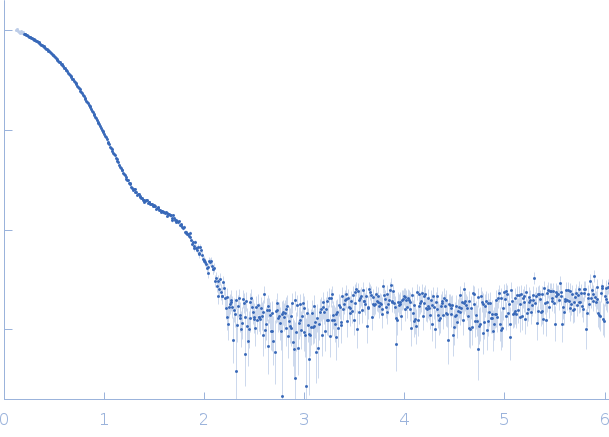
|
| Sample: |
3C-like proteinase from SARS-CoV-2 replicase polyprotein 1a dimer, 68 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 1 mM DTT, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2020 Jun 1
|
|
| RgGuinier |
2.7 |
nm |
| Dmax |
8.8 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
|
|
|
|
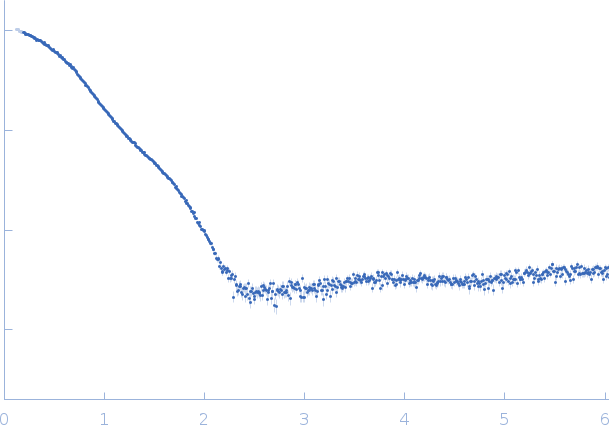
|
| Sample: |
3C-like proteinase from SARS-CoV-2 replicase polyprotein 1a, PT9 mutant monomer, 34 kDa Severe acute respiratory … protein
|
| Buffer: |
50 mM Tris, 1 mM DTT, 1 mM EDTA, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2020 Jun 1
|
|
| RgGuinier |
2.4 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
54 |
nm3 |
|
|