|
|
|
|
|
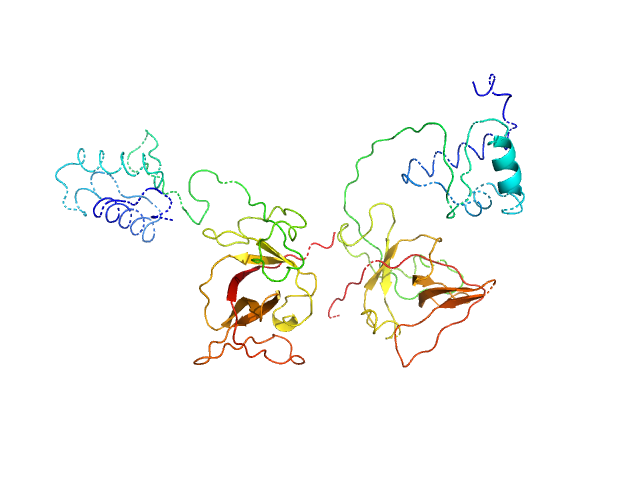
| Sample: |
Bacillus thuringiensis LexA repressor dimer, 47 kDa Bacillus thuringiensis protein
|
| Buffer: |
20 mM Hepes, 300 mM NaCl, 10% glycerol,, pH: 8
|
| Experiment: |
SAXS
data collected at Rigaku BioSAXS-2000, University of British Columbia on 2017 Aug 25
|
Structural Insights into Bacteriophage GIL01 gp7 Inhibition of Host LexA Repressor.
Structure 27(7):1094-1102.e4 (2019)
Caveney NA, Pavlin A, Caballero G, Bahun M, Hodnik V, de Castro L, Fornelos N, Butala M, Strynadka NCJ
|
| RgGuinier |
3.7 |
nm |
| VolumePorod |
110 |
nm3 |
|