Structural variability of EspG chaperones from mycobacterial ESX-1, ESX-3 and ESX-5 type VII secretion systems
Tuukkanen A ,
Freire D,
Chan S,
Arbing M,
Reed R,
Evans T,
Zenkeviciutė G,
Kim J,
Kahng S,
Sawaya M,
Chaton C,
Wilmanns M,
Eisenberg D,
Parret A,
Korotkov K
(2018 Jan 24)
10.1101/250803
Submitted to SASBDB: 2018 Jan 92018 Jan 24
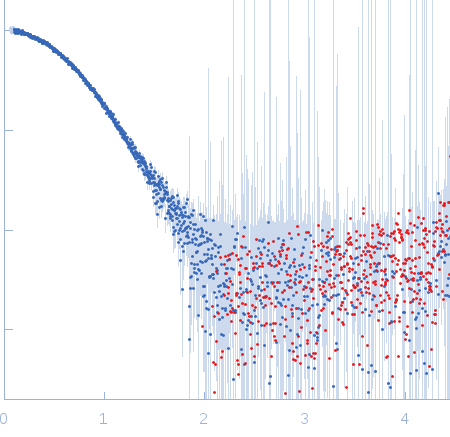
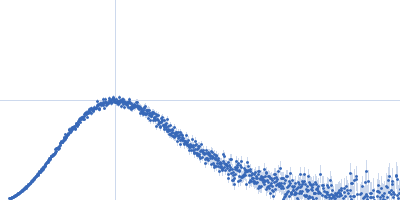
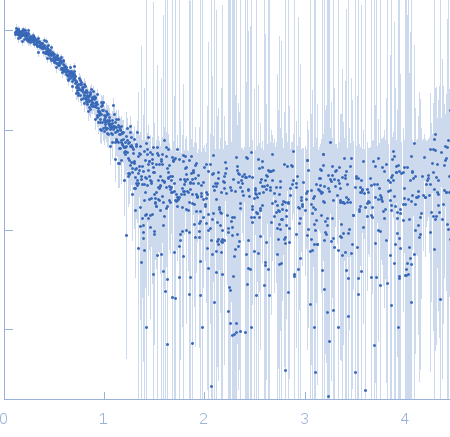
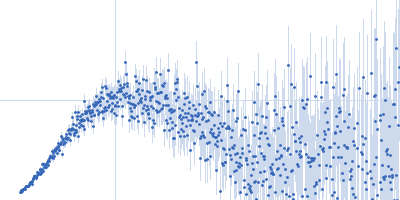
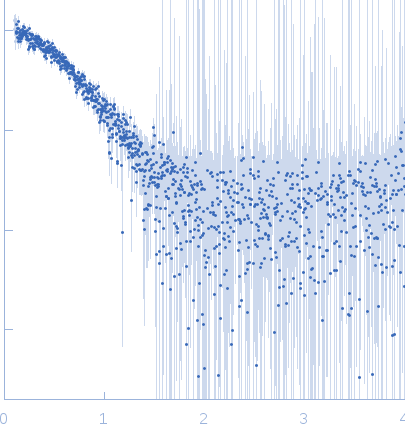
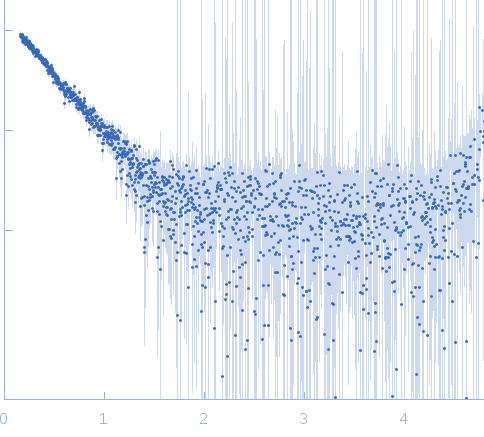
Sample:
EspG3 chaperone from Mycobacterium marinum M monomer, 32 kDa Mycobacterium marinum M protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
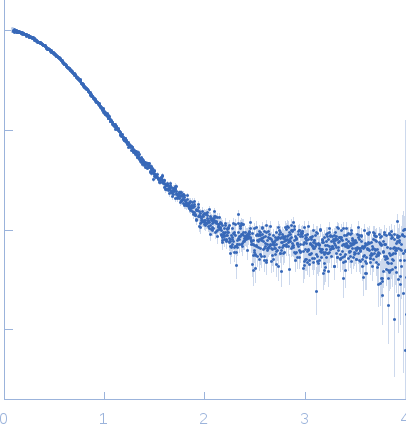
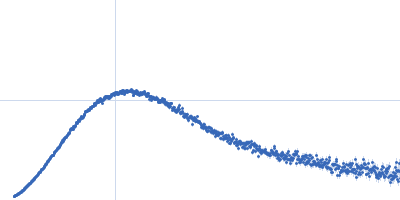
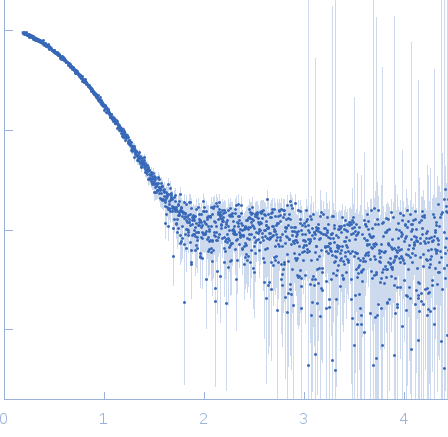
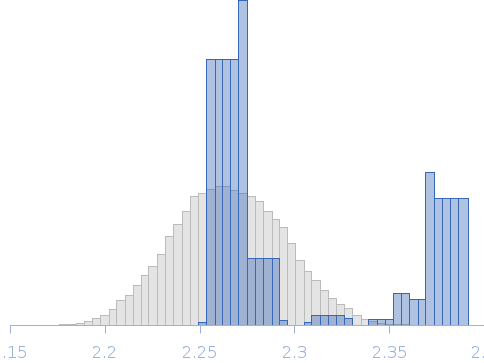
Rg Guinier
2.3
nm
Dmax
8.0
nm
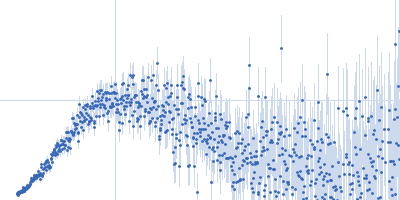
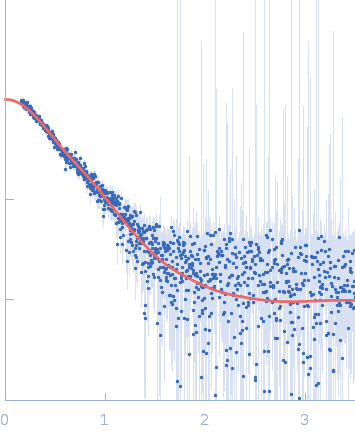
Sample:
EspG1 from Mycobacterium marinum monomer, 30 kDa Mycobacterium marinum protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
Rg Guinier
2.7
nm
Dmax
9.7
nm
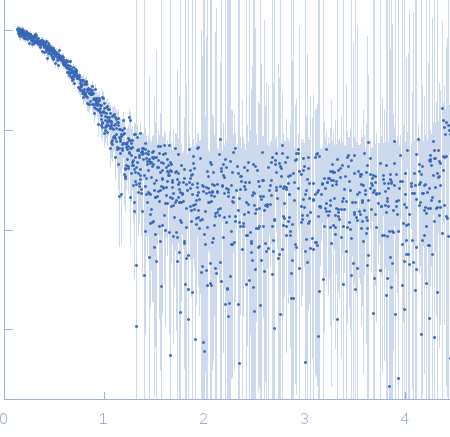
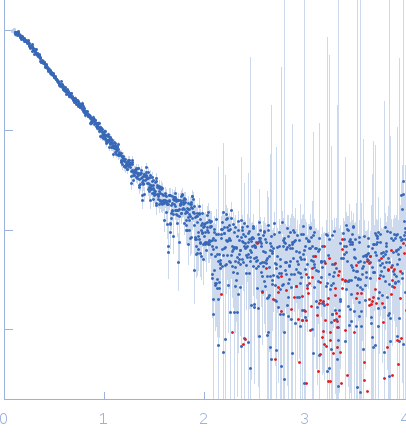
Sample:
EspG3 chaperone from Mycobacterium smegmatis monomer, 32 kDa Mycobacterium smegmatis protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
Rg Guinier
2.5
nm
Dmax
8.6
nm
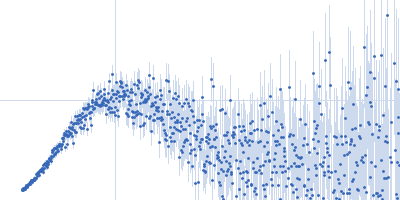
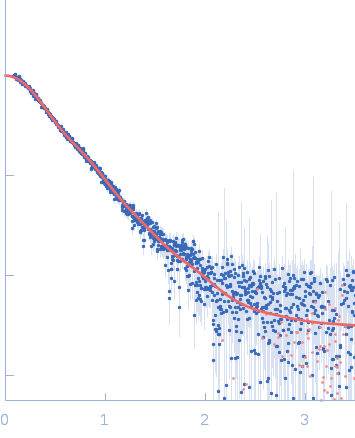
Sample:
EspG3 chaperone from Mycobacterium tuberculosis, 34 kDa Mycobacterium tuberculosis protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
Rg Guinier
2.5
nm
Dmax
9.0
nm
Sample:
EspG3 chaperone from Mycobacterium smegmatis monomer, 32 kDa Mycobacterium smegmatis protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at EMBL P12, PETRA III on 2014 Mar 17
Rg Guinier
2.6
nm
Dmax
9.2
nm
Sample:
EspG5 chaperone from Mycobacterium tuberculosis monomer, 32 kDa Mycobacterium tuberculosis protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at EMBL P12, PETRA III on 2015 Jan 12
Rg Guinier
2.4
nm
Dmax
8.0
nm
Sample:
EspG5 chaperone from Mycobacterium tuberculosis monomer, 32 kDa Mycobacterium tuberculosis proteinMycobacterium tuberculosis proteinMycobacterium tuberculosis protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at EMBL P12, PETRA III on 2015 Feb 10
Rg Guinier
4.0
nm
Dmax
13.0
nm
Sample:
EspG3 chaperone from Mycobacterium tuberculosis, 34 kDa Mycobacterium tuberculosis proteinMycobacterium tuberculosis proteinMycobacterium tuberculosis protein
Buffer:
20 mM HEPES pH 7.5, 150 mM NaCl, pH: 7.5
Experiment:
SAXS
data collected at B21, Diamond Light Source on 2015 Jun 15
Rg Guinier
4.0
nm
Dmax
14.2
nm