|
|
|
|
|
| Sample: |
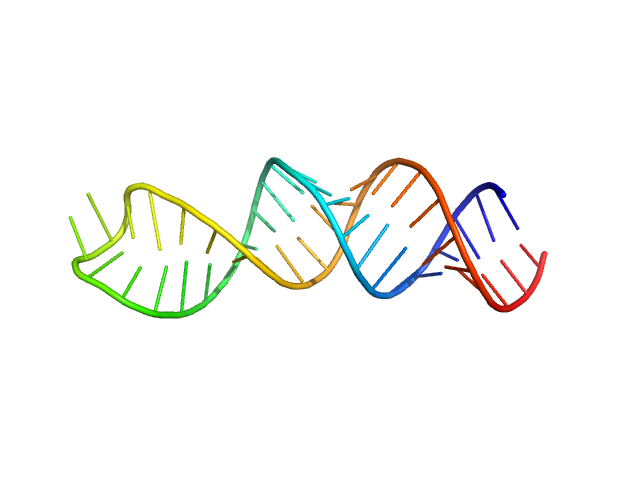
Stem loop 4 in the 5'-genomic end of SARS-CoV-2 monomer, 14 kDa Severe acute respiratory … RNA
|
| Buffer: |
25 mM potassium phosphate, 150 mM KCl, 2 mM TCEP, pH: 6.5
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Nov 22
|
The preference signature of the SARS-CoV-2 Nucleocapsid NTD for its 5'-genomic RNA elements.
Nat Commun 14(1):3331 (2023)
...Dhamotharan K, Jeffries CM, Schlundt A
|
| RgGuinier |
2.0 |
nm |
| Dmax |
7.1 |
nm |
| VolumePorod |
22 |
nm3 |
|