|
|
|
|
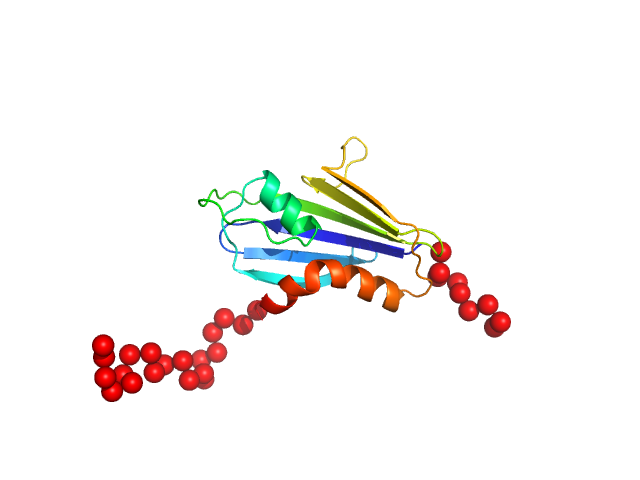
|
| Sample: |
Genome polyprotein monomer, 17 kDa Theiler's murine encephalomyelitis … protein
|
| Buffer: |
20 mM HEPES pH 7.4, 1 M KCl, 2% v/v glycerol, 1 mM DTT, pH:
|
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2021 Apr 26
|
A new protein-dependent riboswitch activates ribosomal frameshifting
(2025)
Betts J, Jeffries C, Passchier T, Kung H, Graham S, Abdelhamid M, Howard J, Craggs T, Graham S, Brierley I, Leake M, Quinn S, Hill C
|
| RgGuinier |
1.9 |
nm |
| Dmax |
7.2 |
nm |
| VolumePorod |
30 |
nm3 |
|