|
Synchrotron SAXS data measured from solutions of the pentameric nucleoplasmin-histone H2A/H2B complex in 20 mM Tris 150mM NaCl, 1mM EDTA, 5mM DTT, pH 8.0, were collected on the BL4-2 beam line at the Stanford Synchrotron Radiation Lightsource (Stanford, USA) using a Rayonix MX225-HE detector (I(s) vs s; s = 4π sin θ/λ, where 2θ is the scattering angle and λ=0.1127 nm). Fifty five successive 1 s data frames were collected immediately after the protein complex eluted from a size-exclusion chromatography (SEC) column. The data were normalized to the intensity of the transmitted beam and radially averaged; the scattering of the solvent-blank (derived from the SEC running solvent) was subtracted and the individual solvent-corrected SAXS profiles were scaled and averaged to produce the final merged SAXS data profile.
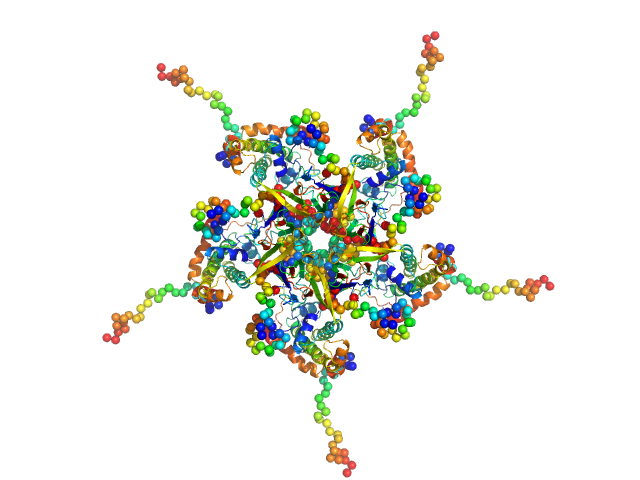
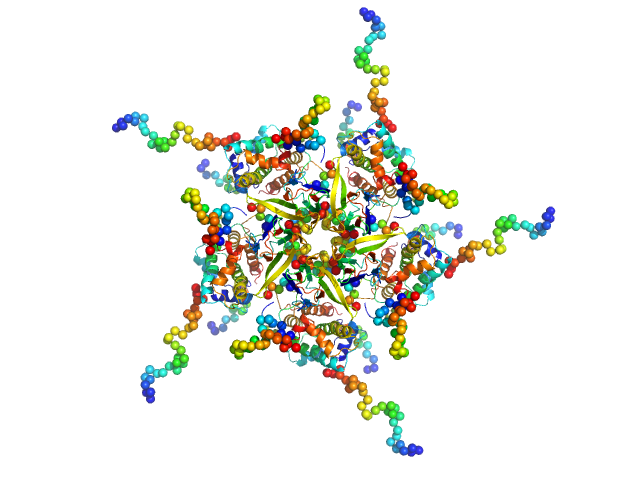
The models displayed for this entry are: Top: ab initio bead model representation (derived from individual reconstructions (example fit shown), aligned, spatially averaged and volume-occupancy corrected): Middle: The best CORAL model using the major A2 site (site #1) of A2:H2A/H2B complex as a starting structure (See Figure 5D in the primary citation). Lacking fragments reconstructed by the program CORAL are shown as spheres. Bottom: The best CORAL model using the minor A2 site (site #2) of A2:H2A/H2B complex as a starting structure (See Figure 5D in the primary citation). Lacking fragments reconstructed by the program CORAL are shown as spheres.
|
|
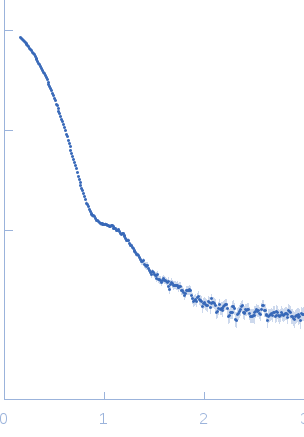 s, nm-1
s, nm-1