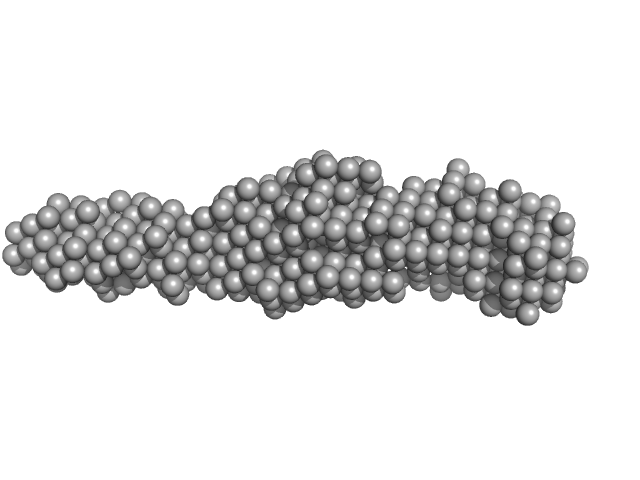
UniProt ID: None (None-None) Collagen like-peptide [GPRG(POG)13]
UniProt ID: Q46085 (811-1021) Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr780Ser, His782Ser, Tyr796Ser and Tyr801Ser)
|
|
|
|
| Sample: |
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD) with Tyr780Ser, His782Ser, Tyr796Ser and Tyr801Ser) monomer, 23 kDa Hathewaya histolytica protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.2 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
34 |
nm3 |
|
|
UniProt ID: Q46085 (811-1021) Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD))
UniProt ID: None (None-None) Collagen like-peptide [GPRG(POG)13]
|
|
|

|
| Sample: |
Collagenase ColH (Polycystic kidney disease domain 2 (PKD2) and Collagen binding domain (CBD)) monomer, 23 kDa Hathewaya histolytica protein
Collagen like-peptide [GPRG(POG)13] trimer, 11 kDa protein
|
| Buffer: |
50 mM HEPES, 100 mM NaCl, 5 mM CaCl2, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2019 Mar 5
|
Elucidating Collagen Degradation Synergy between Col G and Col H from Hathewaya (Clostridium) histolytica and Identifying novel structural features in HPT and REC domains from VarS histidine kinase in V. alginolyticus
University of Arkansas PhD thesis 28030553 (2020)
Perry Caviness
|
| RgGuinier |
3.3 |
nm |
| Dmax |
18.0 |
nm |
| VolumePorod |
35 |
nm3 |
|
|
UniProt ID: P54252 (1-361) Ataxin-3 (polyglutamine protein ataxin-3 (Q13))
|
|
|
|
| Sample: |
Ataxin-3 (polyglutamine protein ataxin-3 (Q13)) monomer, 41 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium phosphate buffer, 2 mM TCEP, 5% glycerol, pH: 6.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 May 24
|
Capturing the Conformational Ensemble of the Mixed Folded Polyglutamine Protein Ataxin-3.
Structure (2020)
Sicorello A, Różycki B, Konarev PV, Svergun DI, Pastore A
|
| RgGuinier |
4.0 |
nm |
| Dmax |
14.1 |
nm |
| VolumePorod |
79 |
nm3 |
|
|
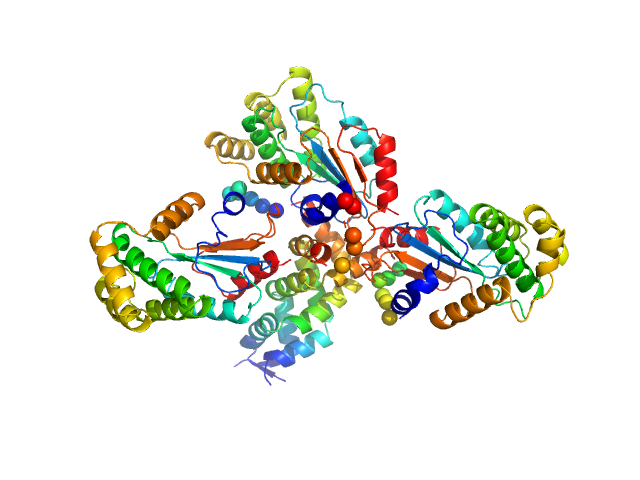
UniProt ID: A0A0H3N7J9 (28-281) Hypothetical exported protein
|
|
|
|
| Sample: |
Hypothetical exported protein trimer, 83 kDa Salmonella typhimurium (strain … protein
|
| Buffer: |
25 mM TRIS, 150 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SAXS/WAXS, Australian Synchrotron on 2019 Aug 14
|
Salmonella enterica BcfH is a trimeric thioredoxin-like bifunctional enzyme with both thiol oxidase and disulfide isomerase activities.
Antioxid Redox Signal (2021)
Subedi P, Paxman JJ, Wang G, Hor L, Hong Y, Verderosa AD, Whitten AE, Panjikar S, Santos-Martin CF, Martin JL, Totsika M, Heras B
|
| RgGuinier |
3.2 |
nm |
| Dmax |
11.5 |
nm |
| VolumePorod |
93 |
nm3 |
|
|
UniProt ID: Q8IKT4 (37-162) Iron-sulfur cluster assembly protein
|
|
|

|
| Sample: |
Iron-sulfur cluster assembly protein trimer, 42 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
50 mM Tris-Cl, 300 mM NaCl, 5% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 May 5
|
[Fe-S] cluster biogenesis and unusual assembly of the ISC scaffold complex in Plasmodium falciparum mitochondrion
Ravishankar Ramachandran
|
| RgGuinier |
5.4 |
nm |
| Dmax |
11.1 |
nm |
| VolumePorod |
177 |
nm3 |
|
|
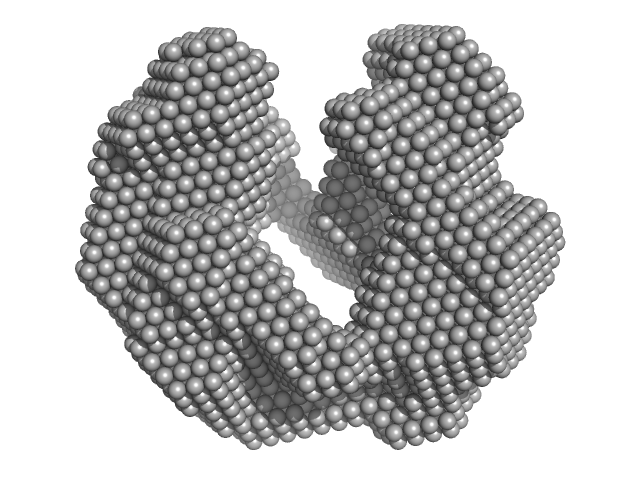
UniProt ID: Q8IKT4 (37-162) Iron-sulfur cluster assembly protein
UniProt ID: Q8IBI5 (104-553) Cysteine desulfurase, putative
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly protein tetramer, 56 kDa Plasmodium falciparum (isolate … protein
Cysteine desulfurase, putative tetramer, 204 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
50 mM Tris-Cl, 300 mM NaCl, 5% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Sep 4
|
[Fe-S] cluster biogenesis and unusual assembly of the ISC scaffold complex in Plasmodium falciparum mitochondrion
Ravishankar Ramachandran
|
| RgGuinier |
6.4 |
nm |
| Dmax |
17.6 |
nm |
| VolumePorod |
1100 |
nm3 |
|
|
UniProt ID: Q8IBI5 (104-553) Cysteine desulfurase, putative
UniProt ID: Q8IKT4 (37-162) Iron-sulfur cluster assembly protein
UniProt ID: Q8IEK7 (1-87) Protein ISD11
|
|
|

|
| Sample: |
Cysteine desulfurase, putative tetramer, 204 kDa Plasmodium falciparum (isolate … protein
Iron-sulfur cluster assembly protein tetramer, 56 kDa Plasmodium falciparum (isolate … protein
Protein ISD11 tetramer, 43 kDa Plasmodium falciparum (isolate … protein
|
| Buffer: |
50 mM Tris-Cl, 300 mM NaCl, 5% Glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at Anton Paar SAXSpace, CSIR-Central Drug Research Institute on 2019 Sep 19
|
[Fe-S] cluster biogenesis and unusual assembly of the ISC scaffold complex in Plasmodium falciparum mitochondrion
Ravishankar Ramachandran
|
| RgGuinier |
7.1 |
nm |
| Dmax |
17.9 |
nm |
| VolumePorod |
950 |
nm3 |
|
|
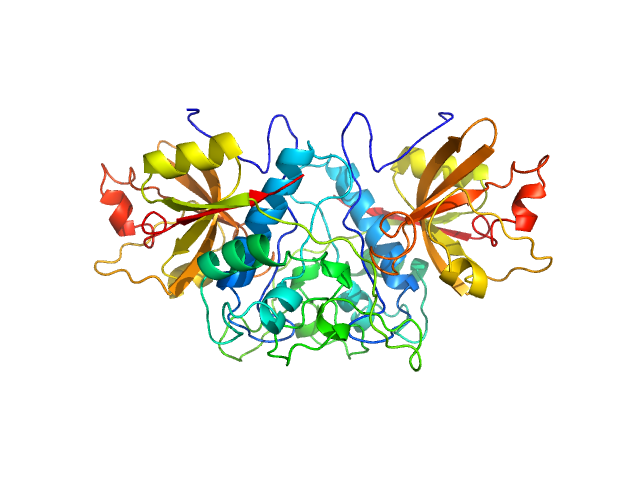
UniProt ID: Q9UBR2 (62-303) Cathepsin Z
|
|
|
|
| Sample: |
Cathepsin Z dimer, 54 kDa Homo sapiens protein
|
| Buffer: |
20 mM sodium acetate, 1 mM EDTA, pH: 5.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2016 Dec 6
|
Human cathepsin X/Z is a biologically active homodimer.
Biochim Biophys Acta Proteins Proteom :140567 (2020)
Dolenc I, Štefe I, Turk D, Taler-Verčič A, Turk B, Turk V, Stoka V
|
| RgGuinier |
2.6 |
nm |
| Dmax |
6.5 |
nm |
| VolumePorod |
89 |
nm3 |
|
|
UniProt ID: Q9DBE0 (1-493) Cysteine sulfinic acid decarboxylase
|
|
|
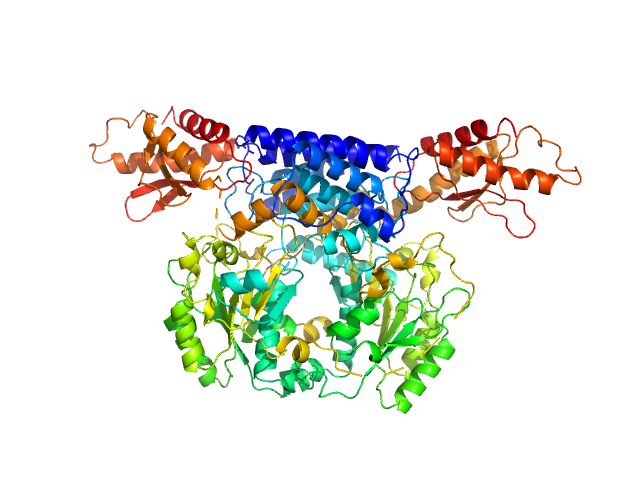
|
| Sample: |
Cysteine sulfinic acid decarboxylase dimer, 117 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 200 mM NaCl, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2018 Feb 4
|
Structure and substrate specificity determinants of the taurine biosynthetic enzyme cysteine sulphinic acid decarboxylase.
J Struct Biol :107674 (2020)
Mahootchi E, Raasakka A, Luan W, Muruganandam G, Loris R, Haavik J, Kursula P
|
| RgGuinier |
3.4 |
nm |
| Dmax |
15.0 |
nm |
| VolumePorod |
198 |
nm3 |
|
|
UniProt ID: Q9C7W1 (1-178) Filaggrin-like protein
|
|
|
|
| Sample: |
Filaggrin-like protein monomer, 20 kDa Arabidopsis thaliana protein
|
| Buffer: |
50 mM HEPES, 150 mM NaCl, 5 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Aug 22
|
Intrinsically disordered plant protein PARCL co-localizes with RNA in phase-separated condensates whose formation can be regulated by mutating the PLD
Journal of Biological Chemistry :102631 (2022)
Ostendorp A, Ostendorp S, Zhou Y, Chaudron Z, Wolffram L, Rombi K, von Pein L, Falke S, Jeffries C, Svergun D, Betzel C, Morris R, Kragler F, Kehr J
|
| RgGuinier |
3.5 |
nm |
| Dmax |
14.0 |
nm |
| VolumePorod |
46 |
nm3 |
|
|