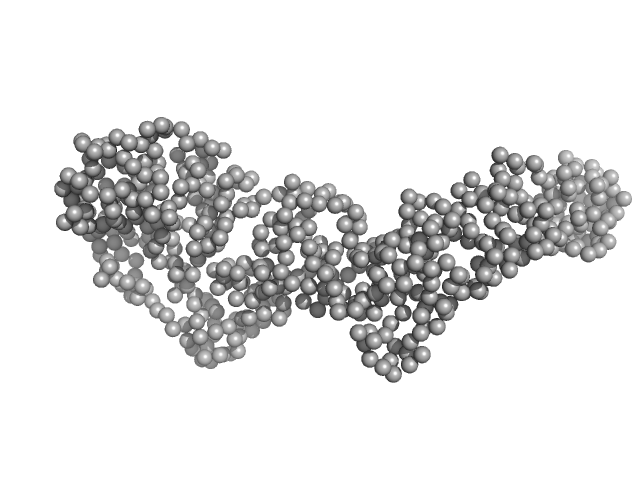
UniProt ID: E6YFW2 (2-558) Bartonella effector protein (Bep) substrate of VirB T4SS
|
|
|
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
| RgGuinier |
3.8 |
nm |
| Dmax |
13.4 |
nm |
| VolumePorod |
87 |
nm3 |
|
|
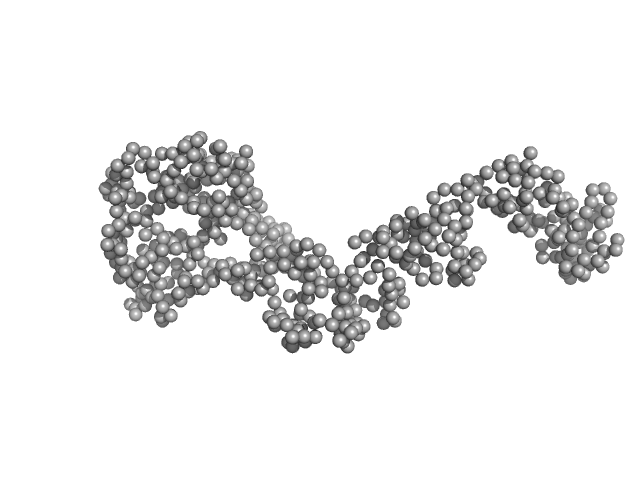
UniProt ID: E6YFW2 (2-558) Bartonella effector protein (Bep) substrate of VirB T4SS
|
|
|
|
| Sample: |
Bartonella effector protein (Bep) substrate of VirB T4SS monomer, 64 kDa Bartonella clarridgeiae (strain … protein
|
| Buffer: |
25 mM Hepes, 300 mM NaCl, 1 mM TCEP, 5% v/v glycerol, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2020 Jun 26
|
Structure and function of Bartonella effector protein 1: target and interdomain interactions
University of Basel PhD thesis 15051 (2023)
Markus Huber, Jens Reiners
|
| RgGuinier |
4.1 |
nm |
| Dmax |
14.4 |
nm |
| VolumePorod |
108 |
nm3 |
|
|
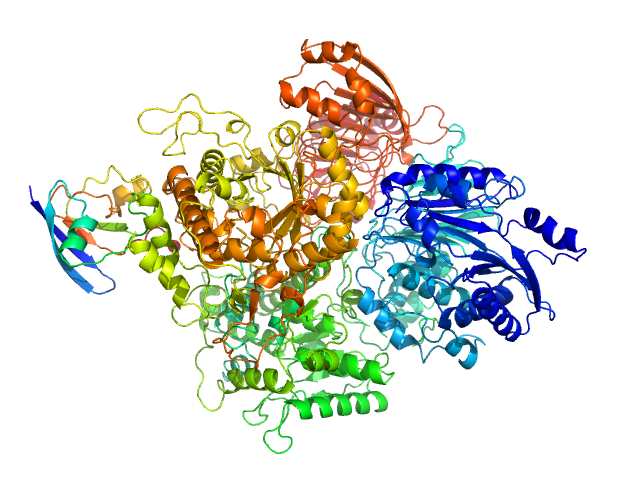
UniProt ID: P55038 (1-1556) Ferredoxin-dependent glutamate synthase 2
UniProt ID: P0A3C8 (1-99) Ferredoxin-1
|
|
|
|
| Sample: |
Ferredoxin-dependent glutamate synthase 2 monomer, 169 kDa Synechocystis sp. (strain … protein
Ferredoxin-1 monomer, 11 kDa Nostoc sp. (strain … protein
|
| Buffer: |
Hepes– KOH buffer, pH: 7.5 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2002 Jun 4
|
The Active Conformation of Glutamate Synthase and its Binding to Ferredoxin
Journal of Molecular Biology 330(1):113-128 (2003)
van den Heuvel R, Svergun D, Petoukhov M, Coda A, Curti B, Ravasio S, Vanoni M, Mattevi A
|
| RgGuinier |
3.6 |
nm |
| Dmax |
12.3 |
nm |
| VolumePorod |
262 |
nm3 |
|
|
UniProt ID: Q08048 (39-728) Hepatocyte growth factor
UniProt ID: P16056 (25-927) Hepatocyte growth factor receptor
|
|
|

|
| Sample: |
Hepatocyte growth factor monomer, 79 kDa Mus musculus protein
Hepatocyte growth factor receptor monomer, 100 kDa Mus musculus protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 9
|
Structural basis of hepatocyte growth factor/scatter factor and MET signalling
Proceedings of the National Academy of Sciences 103(11):4046-4051 (2006)
Gherardi E, Sandin S, Petoukhov M, Finch J, Youles M, Ofverstedt L, Miguel R, Blundell T, Vande Woude G, Skoglund U, Svergun D
|
| RgGuinier |
6.7 |
nm |
| Dmax |
20.0 |
nm |
| VolumePorod |
370 |
nm3 |
|
|
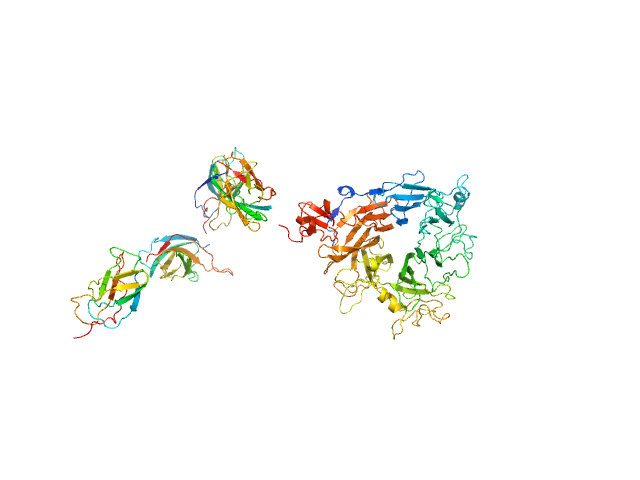
UniProt ID: P16056 (25-927) Hepatocyte growth factor receptor
|
|
|
|
| Sample: |
Hepatocyte growth factor receptor monomer, 100 kDa Mus musculus protein
|
| Buffer: |
50 mM MES, 150 mM NaCl, pH: 6.7 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 Feb 7
|
Structural basis of hepatocyte growth factor/scatter factor and MET signalling
Proceedings of the National Academy of Sciences 103(11):4046-4051 (2006)
Gherardi E, Sandin S, Petoukhov M, Finch J, Youles M, Ofverstedt L, Miguel R, Blundell T, Vande Woude G, Skoglund U, Svergun D
|
| RgGuinier |
4.8 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
190 |
nm3 |
|
|
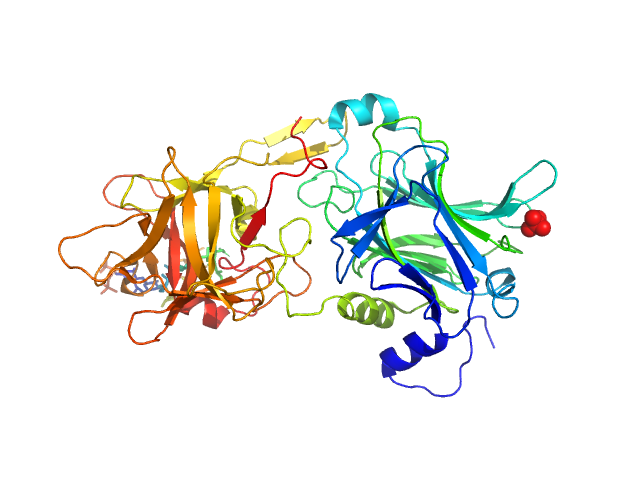
UniProt ID: P04958 (865-1315) Hc fragment of Tetanus toxin
|
|
|
|
| Sample: |
Hc fragment of Tetanus toxin dimer, 103 kDa Clostridium tetani (strain … protein
|
| Buffer: |
50 mM sodium phosphate buffer, 300 mM NaCl, pH: 7.8 |
| Experiment: |
SAXS
data collected at EMBL X33, DORIS III, DESY on 2004 May 14
|
The HC fragment of tetanus toxin forms stable, concentration-dependent dimers via an intermolecular disulphide bond.
J Mol Biol 365(1):123-34 (2007)
Qazi O, Bolgiano B, Crane D, Svergun DI, Konarev PV, Yao ZP, Robinson CV, Brown KA, Fairweather N
|
| RgGuinier |
4.0 |
nm |
| Dmax |
13.2 |
nm |
| VolumePorod |
150 |
nm3 |
|
|
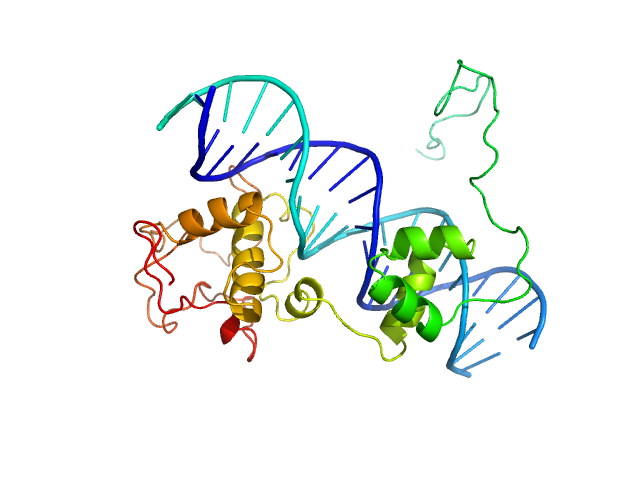
UniProt ID: A0A2D1GKF7 (1-183) Immunity repressor
UniProt ID: None (None-None) 21mer dsDNA
|
|
|
|
| Sample: |
Immunity repressor monomer, 21 kDa Mycobacterium phage TipsytheTRex protein
21mer dsDNA monomer, 13 kDa DNA
|
| Buffer: |
20 mM Tris pH 7.5, 0.5 M NaCl, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at 12.3.1 (SIBYLS), Advanced Light Source (ALS) on 2018 Sep 10
|
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun 13(1):4105 (2022)
McGinnis RJ, Brambley CA, Stamey B, Green WC, Gragg KN, Cafferty ER, Terwilliger TC, Hammel M, Hollis TJ, Miller JM, Gainey MD, Wallen JR
|
| RgGuinier |
2.3 |
nm |
| Dmax |
7.4 |
nm |
| VolumePorod |
41 |
nm3 |
|
|
UniProt ID: P0DN75 (1-132) Iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
| RgGuinier |
4.3 |
nm |
| Dmax |
15.8 |
nm |
|
|
UniProt ID: P0DN75 (2-132) Pigeon iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|
|
| Sample: |
Pigeon iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|
UniProt ID: P0DN75 (2-132) Iron-sulfur cluster assembly 1 homolog, mitochondrial
|
|
|
|
| Sample: |
Iron-sulfur cluster assembly 1 homolog, mitochondrial, 15 kDa Columba livia protein
|
| Buffer: |
20 mM Tris-HCl, 0.15 M NaCl, 10 mM 3-mercapto-1,2-propanediol, pH: 8 |
| Experiment: |
SAXS
data collected at BL-10C, Photon Factory (PF), High Energy Accelerator Research Organization (KEK) on 2020 Nov 30
|
Radiation resistivity of clISCA1
Shigeki Arai
|
|
|