UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2, 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
133 |
nm3 |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2, 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
3.9 |
nm |
| Dmax |
18.5 |
nm |
| VolumePorod |
133 |
nm3 |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2, 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
4.1 |
nm |
| Dmax |
21.5 |
nm |
| VolumePorod |
148 |
nm3 |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2 (R580K)
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2 (R580K), 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
6.1 |
nm |
| Dmax |
32.0 |
nm |
| VolumePorod |
488 |
nm3 |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2 (R580K)
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2 (R580K), 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
5.9 |
nm |
| Dmax |
27.0 |
nm |
| VolumePorod |
381 |
nm3 |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2 (R580K)
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2 (R580K), 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
5.7 |
nm |
| Dmax |
30.0 |
nm |
| VolumePorod |
283 |
nm3 |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2 (R580K)
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2 (R580K), 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
5.3 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
264 |
nm3 |
|
|
UniProt ID: P21980 (1-687) Protein-glutamine gamma-glutamyltransferase 2 (R580K)
|
|
|
|
| Sample: |
Protein-glutamine gamma-glutamyltransferase 2 (R580K), 77 kDa Homo sapiens protein
|
| Buffer: |
20 mM HEPES, 100 mM NaCl, 10% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at ID7A1 BioSAXS / HP-Bio Beamline, Cornell High Energy Synchrotron Source (CHESS) on 2022 Nov 17
|
Distinct conformational states enable transglutaminase 2 to promote cancer cell survival versus cell death.
Commun Biol 7(1):982 (2024)
Aplin C, Zielinski KA, Pabit S, Ogunribido D, Katt WP, Pollack L, Cerione RA, Milano SK
|
| RgGuinier |
5.2 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
243 |
nm3 |
|
|
UniProt ID: J3QMY9 (1-554) Type 2 DNA topoisomerase 6 subunit B-like
|
|
|
|
| Sample: |
Type 2 DNA topoisomerase 6 subunit B-like monomer, 61 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 500 mM KCl, 5% glycerol, 1 mM DTT, pH: 7.5 |
| Experiment: |
SAXS
data collected at SWING, SOLEIL on 2023 Apr 12
|
The TOPOVIBL meiotic DSB formation protein: new insights from its biochemical and structural characterization
Nucleic Acids Research (2024)
Diagouraga B, Tambones I, Carivenc C, Bechara C, Nadal M, de Massy B, le Maire A, Robert T
|
| RgGuinier |
3.7 |
nm |
| Dmax |
15.6 |
nm |
| VolumePorod |
114 |
nm3 |
|
|
UniProt ID: D1C7H4 (1-108) SnoaL-like domain-containing protein
|
|
|
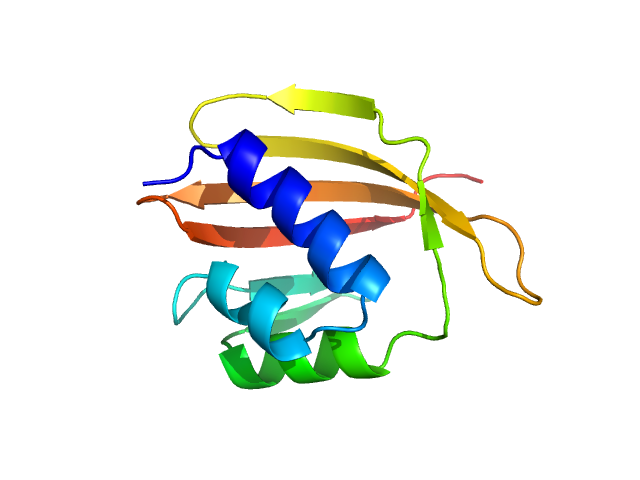
|
| Sample: |
SnoaL-like domain-containing protein monomer, 12 kDa Sphaerobacter thermophilus (strain … protein
|
| Buffer: |
20 mM Tris, 150 mM NaCl, 3% glycerol, pH: 7.6 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2018 Nov 6
|
Elements of the C-terminal tail of a C-terminal domain homolog of the Orange Carotenoid Protein determining xanthophyll uptake from liposomes.
Biochim Biophys Acta Bioenerg 1865(3):149043 (2024)
Likkei K, Moldenhauer M, Tavraz NN, Egorkin NA, Slonimskiy YB, Maksimov EG, Sluchanko NN, Friedrich T
|
| RgGuinier |
1.5 |
nm |
| Dmax |
5.0 |
nm |
| VolumePorod |
18 |
nm3 |
|
|