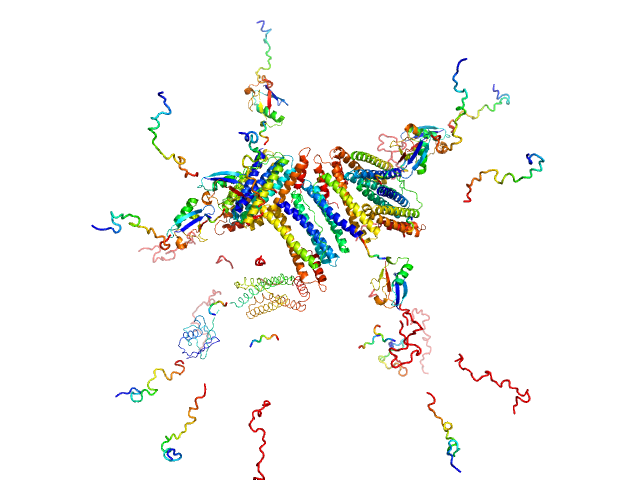
UniProt ID: Q9ZLI1 (5-167) Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion)
|
|
|
|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion) 24-mer, 755 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Jul 30
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
7.6 |
nm |
| Dmax |
25.0 |
nm |
| VolumePorod |
2010 |
nm3 |
|
|
UniProt ID: Q9ZLI1 (5-167) Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion)
|
|
|

|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V, N-terminal His-SUMO fusion) octamer, 252 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
25 mM Tris, 150 mM NaCl, pH: 7.4 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Jul 17
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
5.4 |
nm |
| Dmax |
16.0 |
nm |
| VolumePorod |
618 |
nm3 |
|
|
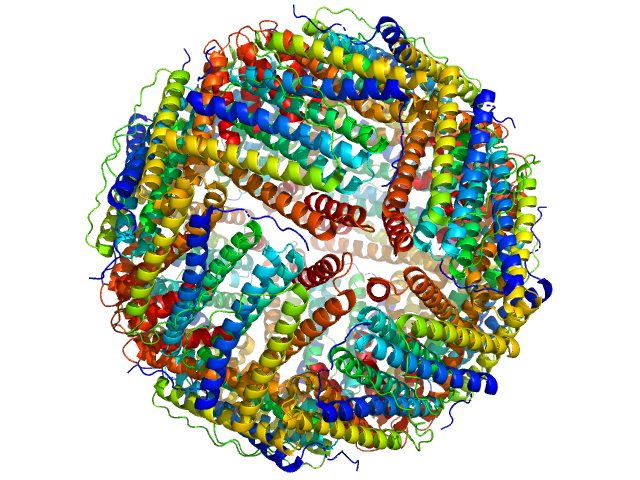
UniProt ID: Q9ZLI1 (5-167) Bacterial non-heme ferritin (N19Q, I59V)
|
|
|
|
| Sample: |
Bacterial non-heme ferritin (N19Q, I59V) 24-mer, 480 kDa Helicobacter pylori (strain … protein
|
| Buffer: |
20 mM Tris, pH: 8 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2022 Dec 14
|
Ferritin-based fusion protein shows octameric deadlock state of self-assembly
Biochemical and Biophysical Research Communications 690:149276 (2024)
Sudarev V, Gette M, Bazhenov S, Tilinova O, Zinovev E, Manukhov I, Kuklin A, Ryzhykau Y, Vlasov A
|
| RgGuinier |
5.6 |
nm |
| Dmax |
14.5 |
nm |
| VolumePorod |
674 |
nm3 |
|
|
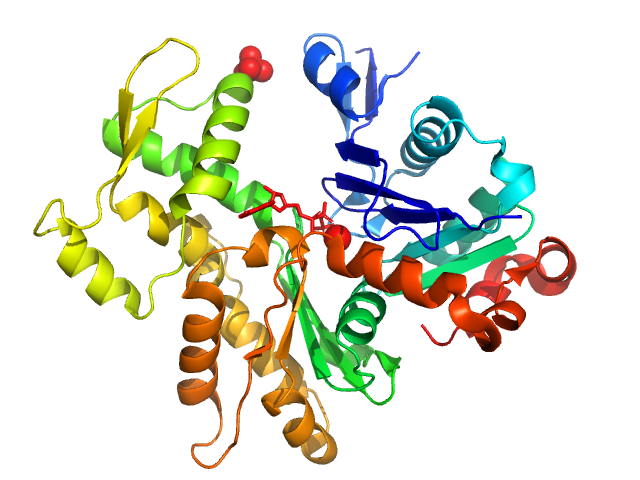
UniProt ID: P68135 (1-377) Actin, alpha skeletal muscle
|
|
|
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 0.1 mM CaCl2, 1 mM NaN3, 0.2 mM ATP, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 1
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
2.9 |
nm |
| Dmax |
8.5 |
nm |
| VolumePorod |
58 |
nm3 |
|
|
UniProt ID: P68135 (1-377) Actin, alpha skeletal muscle
|
|
|

|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 0.1 mM CaCl2, 1 mM NaN3, 1.0 mM ATP, 50 mM KCl, 2 mM MgCl2, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 1
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
15.7 |
nm |
| Dmax |
60.0 |
nm |
| VolumePorod |
4710 |
nm3 |
|
|
UniProt ID: P68135 (1-377) Actin, alpha skeletal muscle
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDTZ4_fit1_model1.png)
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris/Tris-HCl, 2.0 mM EDTA, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Nov 16
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
4.9 |
nm |
| Dmax |
22.0 |
nm |
| VolumePorod |
549 |
nm3 |
|
|
UniProt ID: P68135 (1-377) Actin, alpha skeletal muscle
|
|
|
![OTHER [STATIC IMAGE] model](/media/pdb_file/SASDT25_fit1_model1.png)
|
| Sample: |
Actin, alpha skeletal muscle monomer, 42 kDa Oryctolagus cuniculus protein
|
| Buffer: |
5 mM Tris, 0.1 mM CaCl2, 1 mM NaN3, 0.2 mM ATP, pH: 8.1 |
| Experiment: |
SAXS
data collected at Rigaku MicroMax 007-HF, Moscow Institute of Physics and Technology (MIPT) on 2021 Jul 5
|
Small-angle X-ray scattering structural insights into alternative pathway of actin oligomerization associated with inactivated state
Biochemical and Biophysical Research Communications :149340 (2023)
Ryzhykau Y, Povarova O, Dronova E, Kuklina D, Antifeeva I, Ilyinsky N, Okhrimenko I, Semenov Y, Kuklin A, Ivanovich V, Fonin A, Uversky V, Turoverov K, Kuznetsova I
|
| RgGuinier |
3.1 |
nm |
| Dmax |
12.0 |
nm |
| VolumePorod |
84 |
nm3 |
|
|
UniProt ID: P15116 (160-714) Cadherin-2
|
|
|
|
| Sample: |
Cadherin-2 dimer, 123 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3 mM CaCl2, 0.02% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 1
|
Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell 187(5):1296-1311.e26 (2024)
Tsai YX, Chang NE, Reuter K, Chang HT, Yang TJ, von Bülow S, Sehrawat V, Zerrouki N, Tuffery M, Gecht M, Grothaus IL, Colombi Ciacchi L, Wang YS, Hsu MF, Khoo KH, Hummer G, Hsu SD, Hanus C, Sikora M
|
| RgGuinier |
8.8 |
nm |
| Dmax |
40.0 |
nm |
| VolumePorod |
496 |
nm3 |
|
|
UniProt ID: P15116 (490-702) Cadherin-2
|
|
|
|
| Sample: |
Cadherin-2 monomer, 24 kDa Mus musculus protein
|
| Buffer: |
20 mM HEPES, 150 mM NaCl, 3 mM CaCl2, 0.02% NaN3, pH: 7.5 |
| Experiment: |
SAXS
data collected at 13A, Taiwan Photon Source, NSRRC on 2023 May 1
|
Rapid simulation of glycoprotein structures by grafting and steric exclusion of glycan conformer libraries.
Cell 187(5):1296-1311.e26 (2024)
Tsai YX, Chang NE, Reuter K, Chang HT, Yang TJ, von Bülow S, Sehrawat V, Zerrouki N, Tuffery M, Gecht M, Grothaus IL, Colombi Ciacchi L, Wang YS, Hsu MF, Khoo KH, Hummer G, Hsu SD, Hanus C, Sikora M
|
| RgGuinier |
3.4 |
nm |
| Dmax |
12.6 |
nm |
| VolumePorod |
63 |
nm3 |
|
|
UniProt ID: Q8IYU2 (1-909) E3 ubiquitin-protein ligase HACE1
|
|
|

|
| Sample: |
E3 ubiquitin-protein ligase HACE1 dimer, 205 kDa Homo sapiens protein
|
| Buffer: |
50 mM HEPES, 50 mM NaCl, 5 mM DTT, pH: 8 |
| Experiment: |
SAXS
data collected at EMBL P12, PETRA III on 2023 Apr 21
|
Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1
Nature Structural & Molecular Biology (2024)
Düring J, Wolter M, Toplak J, Torres C, Dybkov O, Fokkens T, Bohnsack K, Urlaub H, Steinchen W, Dienemann C, Lorenz S
|
| RgGuinier |
5.2 |
nm |
| Dmax |
16.4 |
nm |
| VolumePorod |
379 |
nm3 |
|
|